Functional genomics uses genomic data to study gene & protein expression and function on a global scale. Cell-based screening using luciferase reporter assays is a powerful approach for high-throughput gene function discovery as well as drug discovery to assess pathway activity.
Active Motif has taken advantage of the power of reporter assay to develop a complete system specifically optimized for gene regulation experiments. With a genome-wide library of over 30,000 cloned regulatory elements, a highly optimized Renilla luciferase (RenSP) vector system and assay reagents, the LightSwitch™ Luciferase Assay System is the optimal solution for your gene regulation studies. Follow the link for an overview of our wide range of LightSwitch products and services. Information on the LightSwitch technology is provided below under the 'LightSwitch Luciferase Assay Overview' tab.
Advantages of the LightSwitch™ System
- Endogenous human elements (30k total) = Real Biology
Over 30,000 transfection ready promoters, 3´UTRs, and response elements all in the same vector system . - Simple, fast, complete solution
Eliminate the need for assay development and screen immediately with the pre-cloned vectors and tightly optimized assay reagents and protocols. - Quantitative
The novel RenSP luciferase technology allows you to measure promoter activity with unparalleled sensitivity and dynamic range. - Scalable, flexible format options
Convenient 96 to 1,536-well formats available, use for a small number of samples or thousands of assays in a large screen.
White Papers |
Data Sheets |
Product Guides |
Technical Notes |
Posters |
The LightSwitch Luciferase Assay System is a unique collection of over 30,000 regulatory elements that are available in the LightSwitch Promoter and 3´UTR Collections. After selecting a pre-cloned LightSwitch Promoter or 3´UTR Luciferase Reporter Vector, it is transfected into an appropriate cell line. The cells are stimulated, if required, to induce transcription of RenSP luciferase. Gene expression is then quantified using a LightSwitch Assay Kit and a luminometer to measure the amount of luminescence produced.
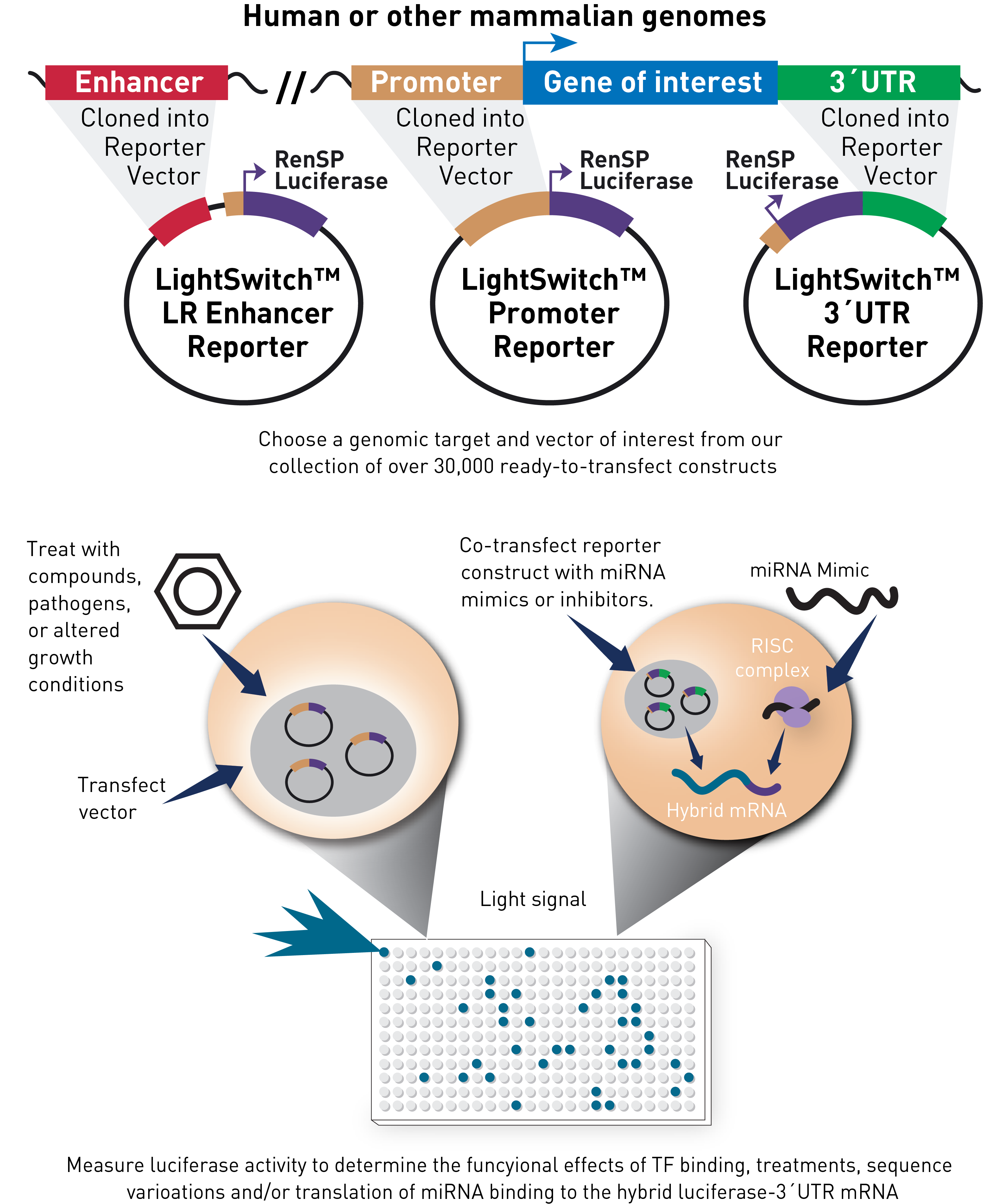
Advantages of the LightSwitch Luciferase Assay System
- Quantitative – Novel RenSP luciferase technology allows you to measure promoter activity with industry-leading sensitivity and dynamic range.
- Simple, fast, complete solution – With pre-cloned LightSwitch Reporter vectors and optimized transfection & assay reagents, you can study regulation of your gene today. No cloning, DNA preparation or optimization is needed, and most studies do not require any internal transfection controls.
- Comprehensive and verified – The genome-wide LightSwitch Reporter Collections are sequence-verified, transfection-ready promoter and 3´UTR reporter vectors.
- Cost-effective – Efficiently screen for activation and/or repression using a multitude of conditions.
RenSP – maximum brightness and minimal background
Why use Renilla luciferase?
Marine luciferases have become popular alternatives to firefly luciferase as a genetic reporter based on assay simplicity, high sensitivity, and a broad linear range of signal that provides greater sensitivity over firefly luciferases.1,2 The Renilla luciferase protein catalyzes oxidation of its coelenterazine substrate in the reaction shown below to produce light at 480 nm, easily read by standard luminometers.3
Inherent advantage of Renilla luciferase
Because marine bioluminescence has evolved independently many times, a variety of luciferases target the same substrate (coelenterazine) yet bear little resemblance to one another.4 Renilla reniformis is a sea pansy that responds to mechanical stimulation by generating a blue-green bioluminescence.4 The small size of its gene and protein (936 bp and 36 kD) and its lack of dependence on ATP provide Renilla luciferase with a distinct advantage over larger, ATP-dependent luciferases like those from fireflies (~1.6 kb and 62 kD).5-8
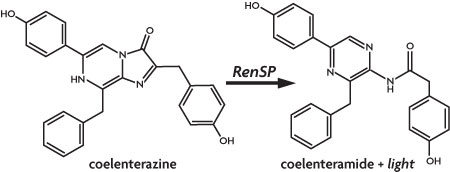
Figure 1: Light is produced from coelenterazine by Renilla luciferase.
Optimizing Renilla for LightSwitch reporter assays – increased enzymatic activity and brightness
We have created an optimized Renilla luminescent reporter gene, called RenSP, by increasing its overall enzymatic activity (light output) and adding a protein destabilization domain to decrease the half-life of the RenSP protein. Starting with a base sequence of the native Renilla gene, we functionally screened thousands of synthetic gene sequence variants that included a variety of predicted improvements. We also removed transcription factor binding sites from the gene sequence as these might confound expression measurements. As a result, we created the RenSP luciferase that is 100% brighter than other humanized versions of Renilla luciferase (Figure 2).9
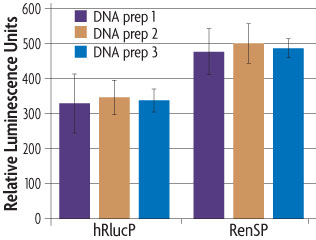
Figure 2: The absolute signal of RenSP is significantly brighter than hRlucP.
To determine the relative brightness of RenSP compared to hRlucP, another humanized form of the Renilla luciferase, the RenSP and hRlucP genes were cloned into separate vectors each containing the human RPL10 promoter. Three independent plasmid purifications were conducted for each vector, and 50 ng of each plasmid was transfected with FuGENE HD in triplicate in human HT1080 cells using a 96-well format. After 24 hours of incubation, 100 µl of LightSwitch Reagent was added to each well and incubated for 30 minutes before being read for 2 seconds on an LmaxII-384 luminometer. These results show that the optimized RenSP luciferase is significantly brighter than hRlucP.
Enhanced degradation rate of RenSP improves assay response
One limitation of reporter gene assays is that the reporter protein can accumulate in the cell; this can delay and dilute the measurable response to stimulation or repression. To eliminate this problem, the RenSP gene has been fused to a protein destabilization domain to reduce the accumulation of reporter protein. The RenSP reporter gene contains a PEST protein degradation sequence from mouse Ornithine Decarboxylase (mODC) that has been shown to increase rates of protein turnover.10-13
The destabilized RenSP luciferase protein has a half-life of approximately 1 hour compared to the ~3 hour half-life of the native luciferase protein, the CAT reporter protein half-life of ~50 hours and the GFP half-life of 25 hours. The RenSP-PEST fusion protein therefore combines the benefits of increased signal with a short half-life reporter to provide a sensitive measure of the induction or repression of reporter gene activity. Figure 3 highlights an example in which signal knock-down of a UTR reporter after addition of a miRNA is more easily detected when the target UTR is fused to a PEST-containing reporter gene.
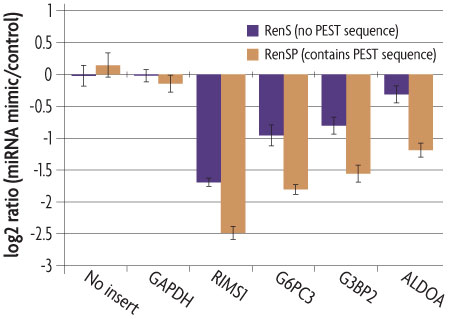
Figure 3: RenSP with PEST increases the knock-down of 3´UTR targets in the presence of mir-122.
The knockdown of 3´UTR-luciferase activity in the presence of mir-122 was measured by co-transfecting a LightSwitch 3´UTR Reporter construct with a synthetic miRNA. DharmaFECT DUO was used to transfect HT1080 cells in triplicate in 96-well format with 100 ng of 3´UTR reporter and 20 nM of mimic or non-targeting control miRNA. After 24 hours of incubation, 100 µl of LightSwitch Assay reagent was added to each well, plates were incubated at room temperature for 30 minutes and read on a LmaxII-384 luminometer. The log2 ratio of the average mimic signal divided by the average signal from the non-targeting control was calculated and shows that the RenSP luciferase with PEST gives a significantly stronger knockdown than RenS without PEST.
LightSwitch™ References
- Lorenz, W., Cormier, M., O’Kane, D., Hua, D., Escher, A., and Szalay, A. (1996) Expression of the Renilla reniformis luciferase gene in mammalian cells. J. Biolumin. Chemilumin. 11:31-37.
- Zhuang, Y., Butler, B., Hawkins, E., Paguio, A., Orr, L., Wood, M., and Wood, K. (2001) A new age of enlightenment. Promega Notes. 79:6-11.
- Hori, K., Wampler, J., Matthews, J., and Cormier, M. (1973) Identification of the product excited states during the chemiluminescent and bioluminescent oxidation of Renilla (sea pansy) luciferin and certain of its analogs. Biochemistry. 12:4463-4468.
- Haddock, S., Moline, M., and Case, J. (2010) Bioluminescence in the sea. Ann. Rev. Mar. Sci. 2:293-343.
- Matthews, J., Hori, K., and Cormier, M. (1977) Purification and properties of Renilla reniformis luciferase. Biochemistry. 16:85-91.
- De Wet, J., Wood, K., Helsinki, D., and DeLuca, M. (1985) Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl. Acad. Sci. 82:7870-7873.
- De Wet, J., Wood, K., DeLuca, M., Helsinki, D., and Subramani, S. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725-737.
- Lorenz, W., McCann, R., Longiaru, M., and Cormier, M. (1991) Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc. Nat. Acad. Sci. 88:4438-4442.
- Zhuang, Y., Butler, B., Hawkins, E., Paguio, A., Orr, L., Wood, M., and Wood, K. (2001) A new age of enlightenment. Promega Notes. 79:6-11.
- Rogers, S., Wells, R., and Rechsteiner, M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 234:364-368.
- Loetscher, P., Pratt, G., and Rechsteiner, M. (1991) The C terminus of mouse ornithine decarboxylase confers rapid degradation on dihydrofolate reductase. Support for the PEST hypothesis. J. Biol. Chem. 266:11213-11220.
- Ghoda, L., Sidney, D., Macrae, M., and Coffino, P. (1992) Structural elements of ornithine decarboxylase required for intracellular degradation and polyamine-dependent regulation. Mol. Cell. Biol. 12:2178-2185.
- pGL4 Luciferase Reporter Vectors Technical Manual. #TM259. Promega Corporation.


