FFPE ChIP-Seq Service Overview
This service is not available at this time.
Epigenetic misregulation is now known to contribute to diseases such as cancer. In cancer, mutations in the enzymes that regulate histone modification deposition and removal are mutated at a high frequency and these mutations result in alterations in global histone modification patterns. Understanding changes in genome-wide histone modification profiles by performing ChIP-Seq assays can provide clues to cellular origins, disease progression, and possibly, patient outcome.
To fully realize the potential of genome-wide histone modification profiling toward advances in disease biology and clinical outcomes, it will be necessary to generate ChIP-Seq data using formalin-fixed, paraffin-embedded (FFPE) patient samples. However, ChIP-Seq is challenging and has traditionally required cell numbers that are much greater than what can be obtained from FFPE sections. To address this limitation, scientists at Active Motif have developed proprietary protocols and workflows to enable generation of high-quality ChIP-Seq data from limited amounts of FFPE tissue.
Let our team of ChIP-Seq experts help run the FFPE ChIP-Seq experiments and do the bioinformatics for you so you can publish faster and in higher impact journals.
The FFPE ChIP-Seq Service Includes:
- Chromatin isolation from FFPE blocks or sections*.
- Immunoprecipitation using a robust ChIP-validated histone modification or transcription factor antibody.
- Library construction for next-generation sequencing.
- Next-generation sequencing using the Illumina platform.
- Comprehensive bioinformatics and delivery of publication-ready figures.
* 5 sections (10 µm) per ChIP-Seq experiment are necessary.
To learn more, please submit an Epigenetic Services Information Request. You can also download Active Motif’s Epigenetic Services Brochure.
FFPE ChIP-Seq Service Data
Active Motif’s FFPE ChIP-Seq Services have been used to generate high-quality ChIP-Seq data from clinical FFPE samples that have been preserved for more than 20 years. Sample types we have successfully processed include tumor FFPE and matched normal FFPE tissue samples from human liver, kidney, colon, and other tissue types. These assays have been performed successfully using ChIP-validated antibodies against multiple histone modifications.
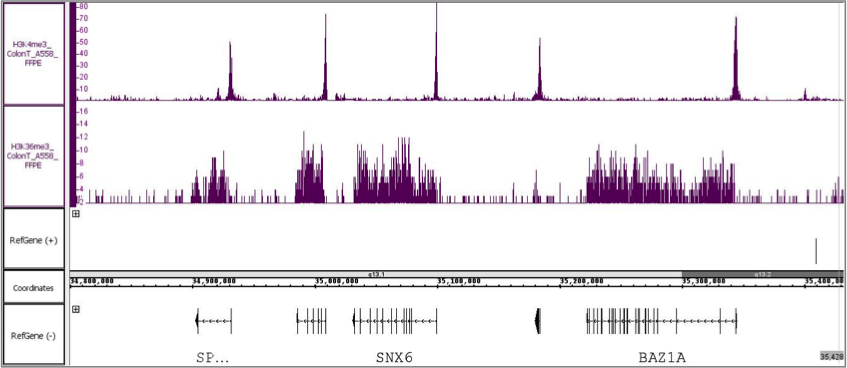
Figure 1: FFPE ChIP-Seq Service performed using a 20-year-old colon tumor FFPE block and antibodies against H3K4me3 and H3K36me3.
Chromatin was extracted from 5 x 10 µm sections derived from a 20-year-old colon tumor FFPE block and ChIP-Seq was performed using antibodies against H3K4me3 and H3K36me3. Each ChIP reaction used approximately 1/3rd of the chromatin preparation. Roughly 500,000 bases of the human genome are depicted in figure 1 showing H3K4me3 occupancy at promoters and H3K36me3 occupancy across gene bodies.
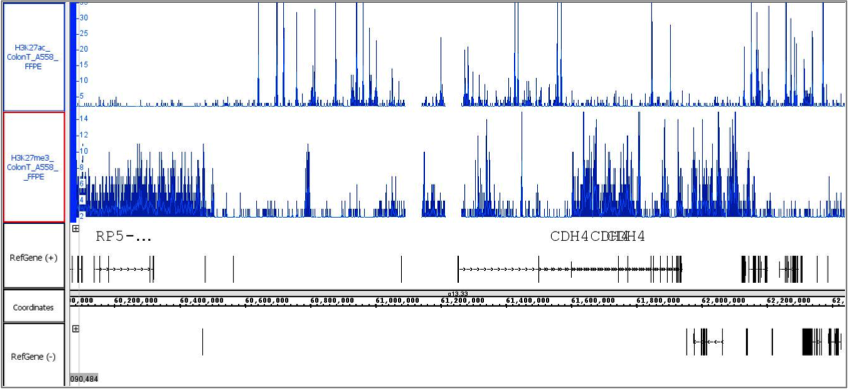
Figure 2: FFPE ChIP-Seq Service performed using 20-year-old colon tumor FFPE block and antibodies against H3K27ac and H3K27me3.
Chromatin was extracted from 5 x 10 µm sections derived from a 20-year-old colon tumor FFPE block and ChIP-Seq was performed using antibodies against H3K27ac and H3K27me3. Each ChIP reaction used only 1/3rd of the chromatin preparation. Roughly 2.4 million bases of the human genome are depicted in figure 2 showing mutually exclusive binding of the enhancer mark H3K27ac and the repressive mark H3K27me3.
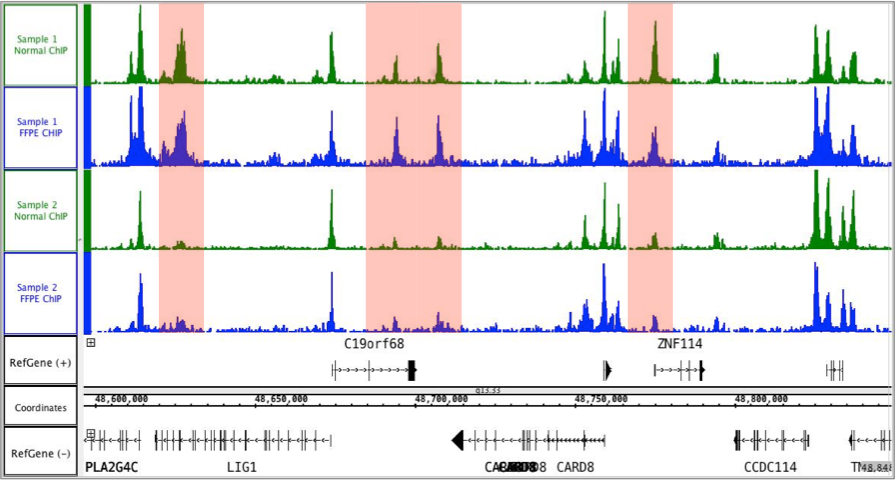
Figure 3: FFPE ChIP-Seq Service performed using 2 human glioblastoma biopsies and an antibody against H3K27ac.
Biopsies were provided as both FFPE prepared samples (blue) and fresh/frozen samples (green). FFPE and fresh/frozen ChIP-Seq data sets were similar. Tumor-specific H3K27ac occupancy was detectable in the FFPE and fresh/frozen data sets (highlighted in red).
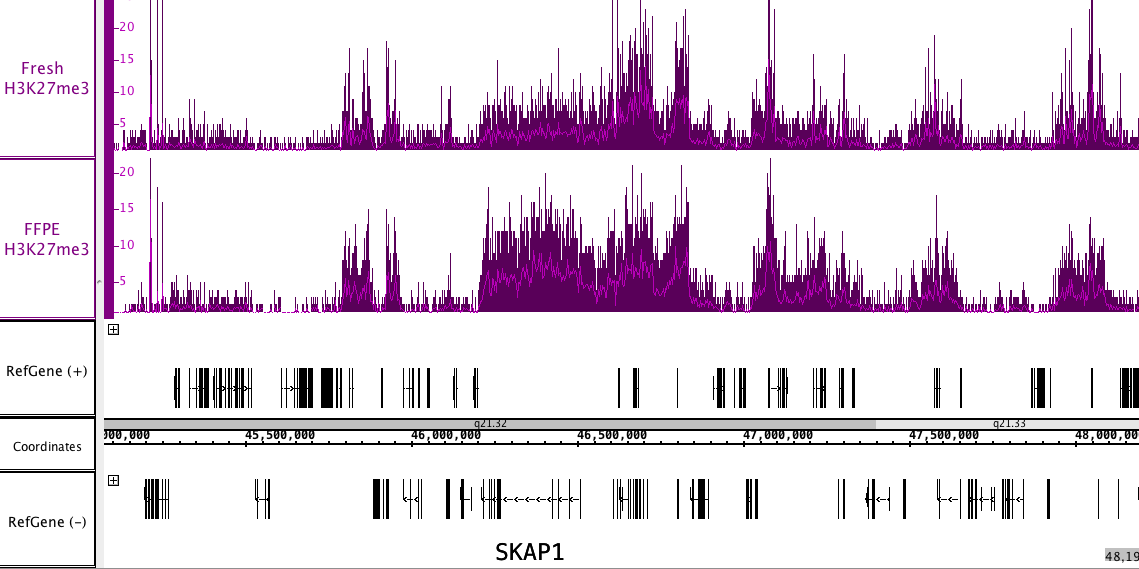
Figure 4: FFPE ChIP-Seq Service performed using a human glioblastoma biopsy and an antibody against H3K27me3.
The biopsy was provided as both a fresh/frozen sample and FFPE tissue block. FFPE and fresh/frozen ChIP-Seq data sets gave similar results. The image shows a typical H3K27me3 occupancy pattern of long stretches of the genome containing the H3K27me3 modification.
FFPE ChIP-Seq Service Documents
| Active Motif Epigenetic Services Brochure |
| FFPE Sample Submission and Requirements Form |
| Epigenetic Services Citations |
FFPE ChIP-Seq Service Sample Submission Portal
Our online sample submission portal allows you to easily upload your service project samples and track your project status. Follow the sample submission instructions in the portal to ensure that all your samples arrive at Active Motif in the best possible condition and properly associated with your project.