<< Back to MOTIFvations Blog Home Page
Reverse Transcribe and Tagment (RT&Tag): A Novel Technique to Explore Chromatin-Associated RNA
May 19, 2023
Table of Contents:
Introduction: The World of Chromatin-associated RNAs
Alongside DNA and protein lies the dynamic world of chromatin-associated RNAs, a less-explored yet critically important domain of the eukaryotic nucleus. Evaluating those chromatin factors that interact with RNAs typically employs complex immunoprecipitation-based protocols with numerous purification steps, where antibodies pull down all RNA associated with an epitope of interest from cell lysates (Gagliardi and Matarazzo). Unfortunately, such immunoprecipitation-based methods generally require large sample inputs and optimization of crosslinking conditions (Gagliardi and Matarazzo and McHugh et al.), which has hampered their more widespread application. These problems prompted the rapid development of more straightforward yet more powerful techniques; now, RNA-focused researchers can turn to a tagmentation-based proximity labeling tool to explore the endogenous chromatin-associated RNA landscape of intact eukaryotic nuclei.
Down to the Basics of RT&Tag
In a recent Nature Methods study, the team of Nadiya Khyzha, Steven Henikoff, and Kami Ahmad (Fred Hutchinson Cancer Center, Seattle, USA) described Reverse Transcribe and Tagment (RT&Tag) as a rapid, low-cost, proximity-labeling tool that facilitates the mapping of chromatin-associated RNAs. This exciting technical advance represents an adaptation of an enzyme-tethering strategy employed to profile protein binding sites in chromatin known as cleavage under targets and tagmentation or CUT&Tag (Kaya-Okur et al.). RT&Tag follows the general framework of CUT&Tag but has been adapted to capture signals from RNA instead of genomic DNA. In brief, RNAs associated with a specific chromatin epitope are targeted by a specific antibody followed by a protein A-Tn5 transposome; subsequently, localized reverse transcription generates RNA/cDNA hybrids that undergo tagmentation by Tn5 transposases for downstream next-generation sequencing.
Proving the Power of RT&Tag
The high efficiency of in-situ antibody tethering and tagmentation by RT&Tag means that each experiment requires significantly fewer input cells (around 100,000) and sequencing reads (around 4–8 million per sample) compared to many conventional RNA immunoprecipitation-based methodologies. Of general importance, RT&Tag forgoes any potential epitope masking from crosslinking, RNA fragmentation, and material loss from numerous purification steps and captures interactions within intact cell nuclei rather than from lysates.
The first example of the utility of RT&Tag employed an antibody against the MSL complex subunit 2 (MSL2) in Drosophila S2 nuclei. This experiment faithfully captured the known interaction of the roX2 non-coding RNA with the dosage compensation complex of proteins (Moran, Niland, and Khalil) but also detected nascent mRNA transcripts across the X chromosome. This proof-of-concept assay proved that RT&Tag could detect trans-acting RNAs in chromatin regulatory complexes and cis-acting RNAs arising from stimulated genes.
Following examples of RT&Tag application detected candidate silencing RNAs and rare unprocessed transcripts from silenced target genes employing an antibody against H3K27me3 (to target Polycomb domains) and post-transcriptionally N6-methyladenosine-modified (m6A) mRNAs with a specific antibody. In the latter example, the authors also revealed that METTL3 binding alone did not support mRNA methylation, suggesting the need for additional factors; in fact, they discovered that promoter pausing of RNA polymerase II characterized genes producing methylated transcripts.
The Future of RT&Tag
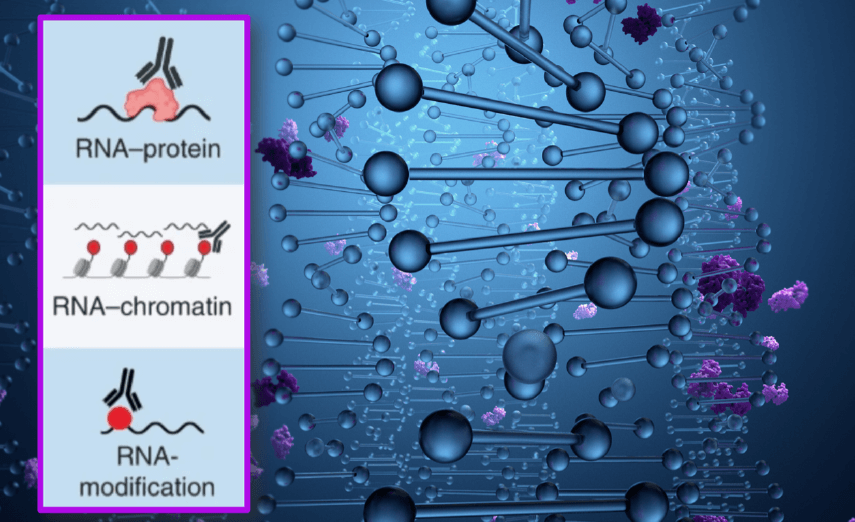
Overall, these experiments highlight the efficacy of RT&Tag when mapping RNA-protein interactions, RNA-chromatin interactions, and RNA modifications within eukaryotic cells; in contrast, more conventional RNA immunoprecipitation techniques require separate protocols for each application. Future developments in RT&Tag may enable the capture of additional functional RNA families (e.g., small-interfering, piwi-interacting, micro, and small-nucleolar RNAs), while subsequent studies may focus on situations in which vanishingly small amounts of biological material have limited in-depth investigations, such as developmental processes.
For more on how RT&Tag can aid the exploration of the chromatin-associated RNA landscape of cells, head over to Nature Methods, October 2022.
About the author

Stuart P. Atkinson, Ph.D.
Stuart was born and grew up in the idyllic town of Lanark (Scotland). He later studied biochemistry at the University of Strathclyde in Glasgow (Scotland) before gaining his Ph.D. in medical oncology; his thesis described the epigenetic regulation of the telomerase gene promoters in cancer cells. Following Post-doctoral stays in Newcastle (England) and Valencia (Spain) where his varied research aims included the exploration of epigenetics in embryonic and induced pluripotent stem cells, Stuart moved into project management and scientific writing/editing where his current interests include polymer chemistry, cancer research, regenerative medicine, and epigenetics. While not glued to his laptop, Stuart enjoys exploring the Spanish mountains and coastlines (and everywhere in between) and the food and drink that it provides!
Contact Stuart on Twitter with any questions
Related Articles
Celebrating 50 Years of RNA Polymerases – the Discovery that Initiated Eukaryotic Transcription Research
September 27, 2019
The discovery of RNA polymerase enzymes was first published in 1969, and the field has come a long way in the last 50 years. This article covers some seminal discoveries in transcription research, the basics of the three mammalian RNA polymerases, and how these enzymes work with other factors to synthesize RNA.
Read More
3’-Digital Gene Expression (3’-DGE) Sequencing: High Through-put at Low Cost
June 14, 2022
The RNA-Seq workflow normally begins with the isolation of RNA and continues with library prep. But RNA can be scary to work with! The scourge of lurking RNases has caused countless restless nights for researchers planning their first (or hundredth!) RNA extraction. Read about an alternative method of library prep, 3'-Digital Gene Expression (3'-DGE) which reduces sequencing depth requirements, the cost of sequencing, and has the potential to deliver good differential gene expression results where conventional RNA-Seq might fail.
Read More
<< Back to MOTIFvations Blog Home Page