Mod Spec® Service の概要
| Name | Cat No. | 価格 (税抜) | |
|---|---|---|---|
| Mod Spec® | 25270 | Get Quote |
Mod Spec® は、質量分析計を使って60種類以上の異なるヒストン修飾の相対量を一度に解析する手法です. Mod Spec® は、エピジェネティックな変化が疾患や薬物処理に応答して生じているかどうかを判断するための出発点になります。多くの修飾変化を一度に見出すことで、予想された変化の確認だけでなく、予期しない重要な変化などを発見できることも期待できます。
日本語チラシはこちら。
ヒストン修飾変化の解析例:
- エピジェネティックな阻害剤への反応
- 正常細胞と罹患細胞の比較
- Knock-out 細胞 や動物細胞
- 薬剤処理した移植サンプル
- ヒト生検サンプル
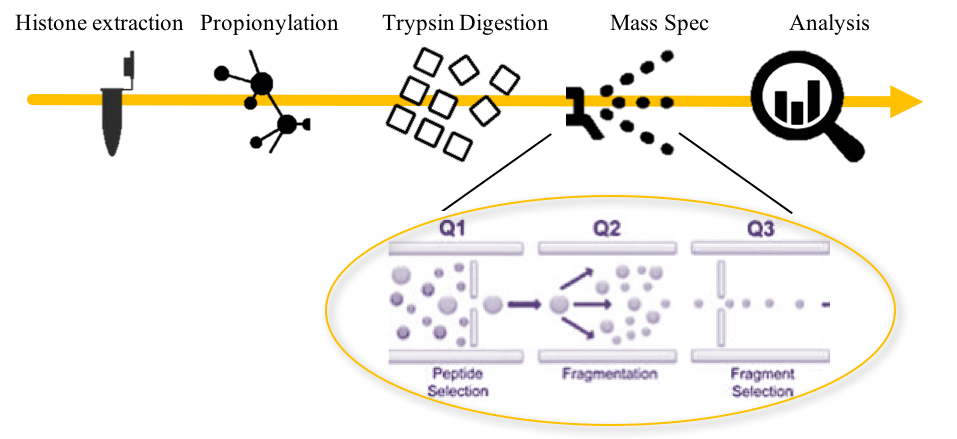
Mod Spec®の仕組み
1. ヒストン抽出: サンプルあたり200~500 万細胞または 25~100 mg の組織が必要です。
2. プロピオニル化: タンパク質の質量分析には、質量分析計に注入する前にタンパク質を小さなペプチドに消化する必要があります。無水プロピオン酸を用いたプロピオニル化は、トリプシンによるリジンの切断を阻害しますが、アルギニンの消化は可能です。これにより得られたペプチドは質量分析に適したサイズになります。
3. トリプシン消化: ヒストンをトリプシンで消化します。
4. 質量分析: トリプル四重極質量分析システムによりサンプルを分析します。
- Q1: 適切な分子量のペプチドが選択されます。
- Q2: ペプチドは衝突チャンバーでさらに断片化されます。
- Q3: 目的のペプチド断片を選択し、質量分析します。
5. 分析: 断片を分析し、サンプル間の変化をグラフ化する。
Histone Modifications Detected by Mod Spec®
^ 複数のH2AアイソフォームがH2Aの修飾シグナルに寄与している可能性があります。
* H3R2me2とH3K4me2/3は互いに排他的な修飾です。 H3R2のメチル化はH3K4のメチル化を妨げるため、H3K4修飾が検出されるのはH3R2が未修飾の場合のみです。 詳しくは、以下の文献をご参照ください:Nature. 2007 Oct 18; 449(7164):933-7.
♦ K27およびK36近傍のアミノ酸配列はH3.1とH3.2で保存されているため、H3.1の結果にはH3.2の修飾も含まれる可能性があります。
What our customers are saying about us:
"While working on the molecular mechanism of an epigenetic drug, we outsourced Mod Spec® to Active Motif in order to get a broader overview of the drug-induced changes of histone modifications. Overall, we were very pleased with the quality of the service, the kept timeline and last but not least, the fair price. We found surprising things that were not on our radar before."
Matthias Lauth, PhD
Phillips University
Marburg, Germany
View complete list of testimonials >
Mod Spec® 受託解析サービスのデータ
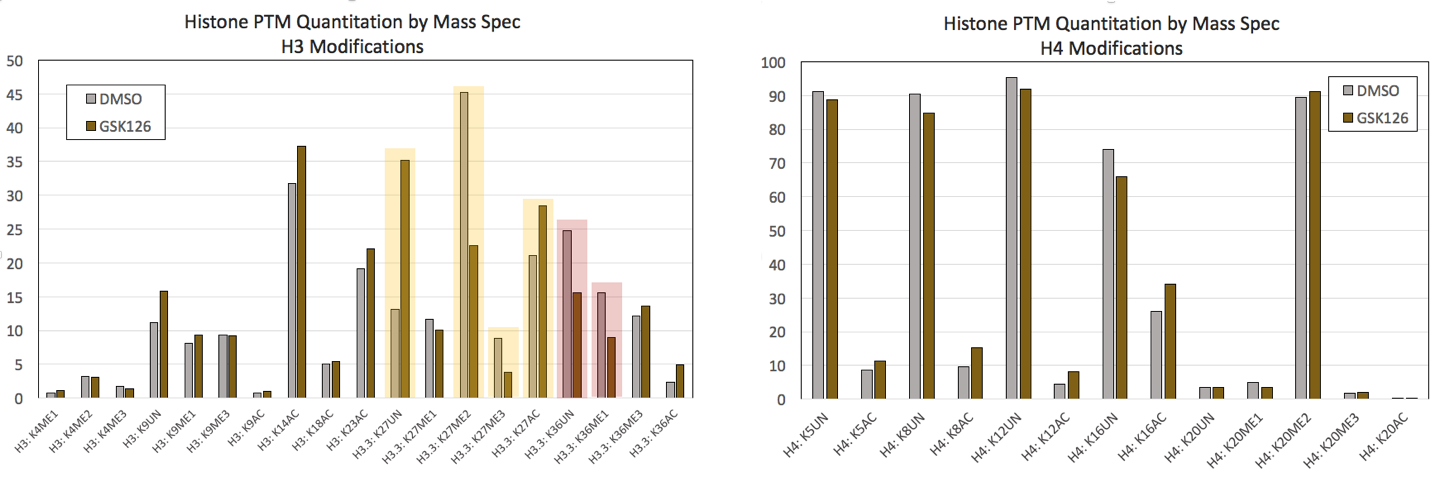
図1:コントロールおよび0.5 µM GSK-126で7日間処理したHeLa細胞のMod Spec®のデータ(画像をクリックすると拡大します)。(Click image to enlarge)
GSK126はEZH2のメチルトランスフェラーゼ活性を阻害するため、その投与は、H3K27me2、およびH3K27me3をグローバルに減少させることが知られている。アクティブ・モティフのMod Spec®アッセイによるH3修飾の一部を示す(左)。未修飾、およびアセチル化されたH3K27(黄色で強調)は増加に加え、H3K27メチル化の有意な減少が確認された。 H3K36の変化も検出された(赤で強調)。GSK126処理に対して有意な変化を示さないH4修飾の一部(右)。
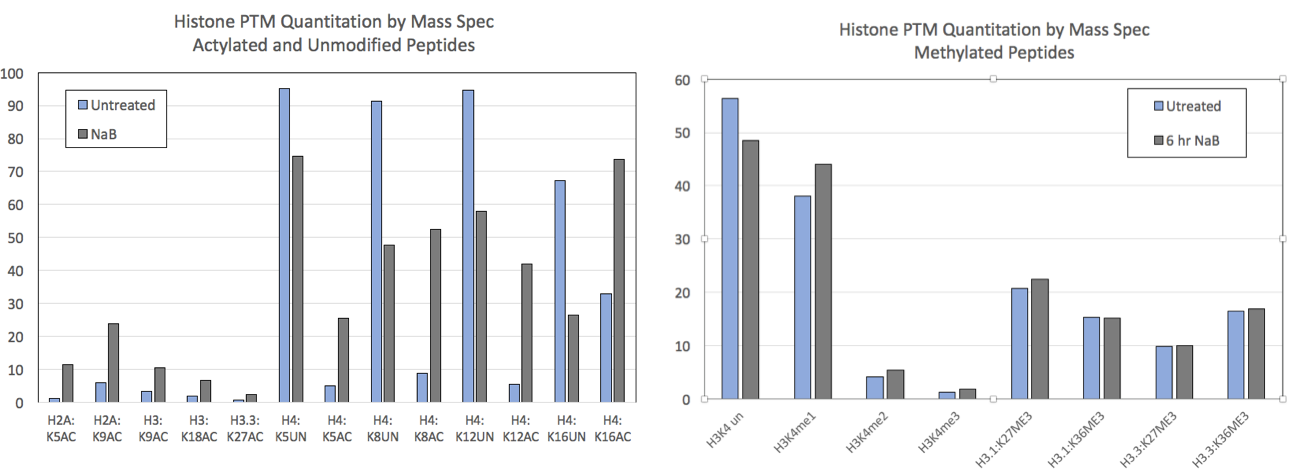
図2:コントロールおよび5mM sodium butyrate (NaB)で6時間処理したHEK293細胞のMod Spec®のデータ。
NaBは代表的なHDAC阻害剤であり、投与によりヒストンのアセチル化レベルが上昇すると予想される。アセチル化される部位と対応する未修飾部位のMod Spec®データを示す(左)。アセチル化されうる全ての部位において、NaB処理後にアセチル化が増加している。ここに示されている未修飾ペプチドは、NaB処理により減少しており、このことは、これらの部位が内因性のHDACにより脱アセチル化されることを示唆している。ヒストンのメチル化は、多くの部位においてNaB処理による影響を受けない(右)。
Mod Spec® Service 使用文献
Search our database of customer publications that have used our Mod Spec® services.
Mod Spec® 受託解析サービスの資料
Mod Spec® Service Sample Submissionポータル
オンラインSample Submissionポータルは、解析したいサンプルの情報をアクティブ・モティフの担当者と共有し、依頼者には解析の進捗をご覧いただけるサイトです。 解析を依頼するために必要な連絡先、サンプルの情報、また解析の内容を記載していただきます。記載の方法や、サンプル提出方法について、ご不明な点がありましたら「japantech@activemotif.com」までご連絡ください。