<< Back to MOTIFvations Blog Home Page
Celebrating 50 Years of RNA Polymerases – the Discovery that Initiated Eukaryotic Transcription Research
September 27, 2019
The “central dogma” of molecular biology has been drilled into hundreds of thousands of biologists and high school students alike since the concept was first proposed in 1957. Scientists in the late 1950s were armed with the knowledge of the three major biological molecules (DNA, RNA, and proteins), but it was Francis Crick who envisioned the directional flow of information between them.
Following Crick’s hypothesis, the first DNA-directed RNA polymerase activity was detected in both E. coli and rat liver nuclei in 1960. One year later, in 1961, mRNA was identified as the intermediate between DNA and proteins, and tRNA was isolated as the adaptor molecule linking mRNA to proteins. The rRNA molecule was classified shortly thereafter, in 1962.
It was clear that these single-stranded RNA transcripts played a major role in relaying the information contained in the genetic code, but it still wasn’t clear how they were formed. This effort was hindered by the inability to study the generation of specific RNAs.
In his recently published historical review of the discovery of RNA polymerase enzymes, Robert Roeder recalls his desire “to [get] to the heart of the matter—the RNA polymerase”. Accordingly, Roeder’s work led to the landmark 1969 discovery of three chromatographically distinct mammalian RNA polymerases, and subsequent groundbreaking research on their unique structures and functions. These seminal discoveries are discussed in more detail below, as well as current models of the three mammalian RNA polymerases, and how they work in concert to produce the three major classes of RNA.
How was Gene Expression Studied in the Early Days?
The elucidation of the lac operon regulation in E. coli by François Jacob and Jacques Monod in 1961 might be considered the inaugural event in the field of gene regulation. These discoveries were made well before the advent of cloning, PCR, and mail-order sequencing, so these gene regulation pioneers relied on complementation assays to elucidate the mechanism by which related genes coordinate their expression with other regulatory elements and respond to external environmental factors.
Work in E. coli and other prokaryotic systems dominated the early gene expression research studies. Prokaryotes held many advantages for experimental design, including simplicity (smaller genomes and lack of nucleus), fast replication rates, and of course, low cost. Although RNA polymerase activity was reported in prokaryotes (in E. coli) and eukaryotes (in rat liver nuclei) simultaneously, much of the early work demonstrating the DNA-dependence of RNA polymerase was done in E. coli.
On the eukaryotic side of research, work was focused on the simple eukaryotic budding yeast S. cerevisiae for many of the same reasons that many scientists worked with prokaryotes. For gene-related studies in particular, rat liver cells and/or sea urchin embryos were popular, since techniques had been previously developed to extract and isolate the nuclei from these cell types.
Three Different Mammalian RNA Polymerase Enzymes
In the 50 years since the discovery of the three mammalian RNA polymerases, we’ve come to appreciate their diverse functions, specific gene targets, and unique transcription products. Today, RNA polymerase II (Pol II) is perhaps the most well recognized of the group. Comprised of 12 subunits, it is responsible for the production of messenger RNA (mRNA), which is in turn translated into a polypeptide—the “central dogma” polymerase. RNA polymerase I (Pol I) has 14 subunits and is dedicated solely to the production of rRNA required for the large ribosomal subunit which, together with the small ribosomal subunit and a variety of ribosomal proteins, forms the ribosome. The 17 subunit RNA polymerase III (Pol III) is the most diverse in its transcription products, which include tRNA, the small ribosomal subunit rRNAs, and other small RNAs like spliceosomal RNAs, snRNAs, and miRNAs.
Discovery of the RNA Polymerases
A paper published in 1964 on the differential RNA polymerase enzymatic activities at different salt and ion concentrations was a clue that there may be distinct enzymes/polymerases responsible for the production of different RNAs, but the initial discovery of the three distinct mammalian RNA polymerases came from Robert Roeder in the lab of William Rutter in 1969.
Roeder developed a novel method for solubilizing the RNA polymerase enzymes while removing the contaminating DNA and histone components—a barrier that had thwarted previous attempts at purification. Using enzyme preparations from sea urchin embryos and rat liver nuclei, Roeder was able to identify three distinct enzymes eluted during ion-exchange chromatography. Roeder performed further experiments measuring specific activity in the nucleus and nucleolus cell fractions, confirming that Pol I is in the nucleolus and Pol II and Pol III are found in the nucleoplasm.
The next logical step was to determine the functions of the individual polymerases. Conveniently, each of the RNA polymerases is differentially sensitive to the toxin alpha-amanitin: Pol I is unaffected, Pol II is inhibited at low concentrations, and Pol III is inhibited at high concentrations.
This key piece of information allowed several groups to dissect the RNA polymerase transcription products, ultimately determining that Pol I is responsible for rRNA 45S synthesis, Pol II for the production of mRNA, and Pol III was shown to be responsible for the transcription of tRNA and small ribosomal subunit rRNA.
Noting strong evolutionary conservation between prokaryotic and eukaryotic polymerases, as well as among the mammalian RNA polymerases, it is hypothesized that duplicated copies of the prokaryotic RNA polymerase evolved to more efficiently transcribe specific types of RNA.
RNA Polymerase Sequences & Structures
The amino acid sequences of the RNA polymerases were not identified until the era of cloning and genomic sequencing in the 1990s. This was no small feat considering the number of subunits for each polymerase: 14, 12 and 17 subunits for Pol I, II, and III, respectively.
In the years that followed the cloning and sequencing of the gene encoding the RNA polymerase enzymes, researchers began to turn to structural studies to try to solve their three-dimensional structures. Roger Kornberg and colleagues obtained a high-resolution crystal structure of Pol II, which they published in 2001, and the structures of Pol I and Pol III soon followed, further revealing conserved structures, structural insights, and earning Kornberg the 2006 Nobel Prize in Chemistry.
The Role of Transcription Factors & Other Transcriptional Co-Factors in Gene Expression
The singular prokaryotic RNA polymerase, with its lone transcription factor, is in stark contrast to the complexity of the eukaryotic RNA polymerases. It was quickly determined that an isolated RNA polymerase and a target gene is not enough to initiate transcription. But take the same two components and put them in an RNA polymerase-free cell extract, and transcription is initiated.
Thus, the next logical step was to establish what components were required for initiation of transcription, and determine if these requirements vary between the three polymerases.
General Transcription Factors
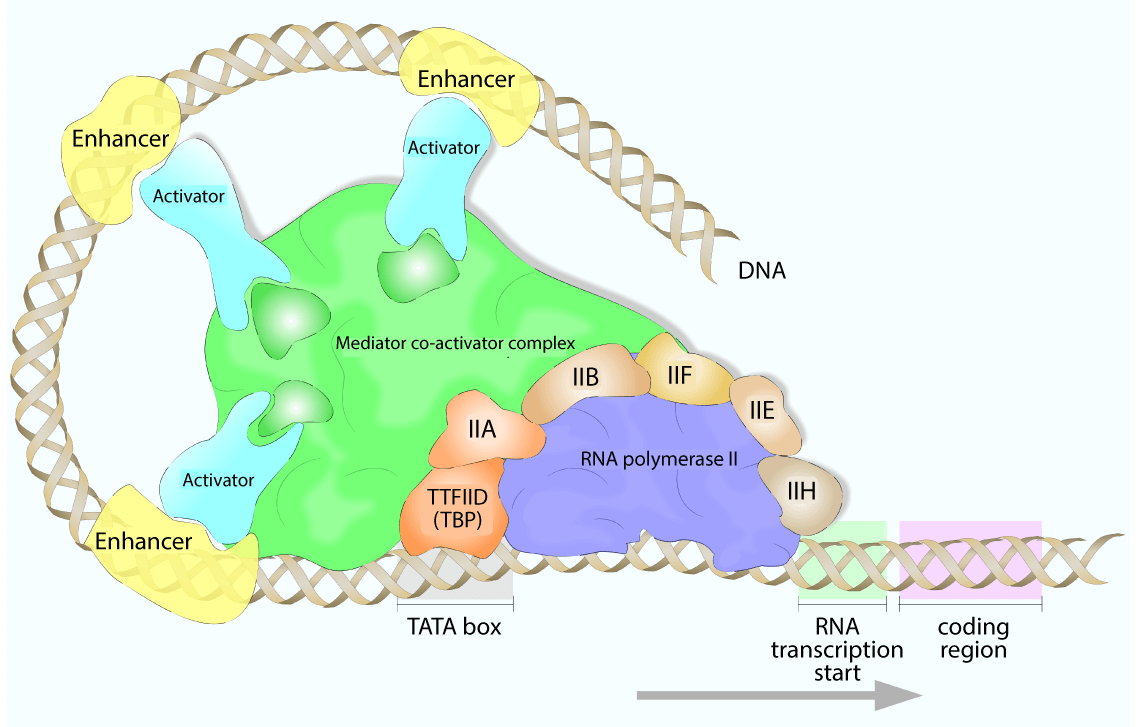
For all the RNA polymerases, the assembly of a preinitiation complex (PIC) is required for successful transcription. RNA polymerase I, II, and III each have different pathways associated with PIC assembly, but have many shared components and overlapping mechanisms.
The minimum requirement for PIC formation and transcription initiation is RNA polymerase plus several general transcription factors (GTFs). Biochemical legwork elucidated the varying polymerase/transcription factor pairings, which are listed in the table below:
| Enzyme | Transcription Product | Minimum Required Initiation Factors |
|---|---|---|
| RNA Pol I (14 subunits) |
rRNA 45S | SL1*/TIF1B/core factor TIF1A/RRN3 UBF/UAF |
| RNA Pol II (12 subunits) |
mRNA snRNA miRNA |
TFIIA TFIIB TFIID* TFIIE TFIIF TFIIH |
| RNA Pol III (17 subunits) |
tRNA rRNA 5S small RNAs |
TFIIIC TFIIIB* |
*contain the TATA-binding protein
For each polymerase, one of the GTFs binds the TATA-box, constraining the transcription start site to the specified promoter. Additional transcription factors are then recruited, which often have secondary enzymatic activities, such as helicases, kinases, or ATPases. Additionally, RNA polymerase itself is recruited to the growing PIC by the GTFs.
For RNA polymerase II, an additional required factor was identified: the Mediator complex. The human Mediator complex has 30 subunits, spanning contact points with transcription factors in the PIC and with Poll II. In the most simplistic view, the Mediator complex acts as a “bridge” between transcription factors and Pol II and stabilizes PIC formation. In this way, it relays signals from transcription factors to the polymerase machinery to regulate the transcriptional activity.
More recently, the Mediator complex has been shown to function in epigenetic regulation, chromatin remodeling, and super-enhancer formation, making it a crucial regulatory factor for RNA polymerase II.
Other Transcriptional Co-Factors
Although the general transcription factors and RNA polymerase enzymes comprise the minimum components of transcription initiation, these factors alone generally lead to low basal transcription activity.
There can be up to 100 proteins in the active RNA polymerase complex, functioning to stabilize, promote, and regulate transcription. Generic activators may bind enhancer regions and interact with the RNA polymerase machinery and increase its affinity for the promoter region, while repressors bind to silencer regions to block transcription.
Coactivators, on the other hand, do not directly interact with the target gene, but instead regulate transcription via interactions with transcription factors or through chromatin remodeling.
Since cell-specific genes can be transcribed by the general RNA polymerase II machinery, cells must have regulatory mechanisms to prevent promiscuous and continuous transcription of all cellular genes. Many regulatory elements are bound by cell- or gene-specific activators or repressors, such as TFIIIA, the first described gene-specific activator, and the OCA-B coactivator, which regulates B-cell specific gene transcription.
Epigenetic Regulation of Transcription Initiation
Another layer of regulation exists at the target genes themselves, in the form of epigenetic modifications and chromatin accessibility. Transcriptional coactivators can act as chromatin remodelers, usually through histone acetyltransferase (HAT) activity, which works by reducing the affinity of DNA for histones, loosening the chromatin structure and allowing transcriptional machinery to bind.
A well-characterized example is the p300 coactivator, which functions as a HAT, and additionally binds other transcription factors to recruit RNA polymerase machinery. p300 has been shown to interact with more than 50 other proteins, allowing for the regulation of transcription in critical cellular processes such as proliferation, differentiation, and apoptosis.
RNA Polymerase II Post-Translational Modifications (PTMs)
Although it seems like 6 initiation factors, a 26 subunit Mediator complex, and many more activators, repressors, and chromatin remodelers would be enough regulation, there are several post-translational modifications (PTMs) of the Pol II enzyme itself that additionally control transcription.
The largest subunit of Pol II has a carboxy-terminal domain (CTD) comprised of 52 repeats of the following consensus sequence: Y1S2P3T4S5P6S7. This repeated heptamer undergoes two major phosphorylation events after Pol II is recruited to the PIC.
For Pol II to escape the promoter, serine 5 must be phosphorylated by the transcription factor TFIIH/CDK7. This is turn recruits the complex needed for 5’ capping of newly synthesized mRNAs. Secondly, serine 2 is phosphorylated by the kinase P-TEFb/CDK9 to promote stable elongation of the nascent mRNA via dissociation of a negative elongation factor complex.
As elongation continues and Pol II approaches the end of the gene, a variety of phosphatases dephosphorylate these same positions such that Pol II can initiate another round of transcription. It should be noted that not all of the 52 available Ser5 or Ser2 sites are phosphorylated, and the dynamic interplay between these two sites provides an additional layer of regulation.
What’s Next for RNA Polymerases and Eukaryotic Transcription Research
The last 50 years have seen a rapid expansion of knowledge regarding the structure and function of the eukaryotic RNA polymerases, as well as other factors that influence transcription. As it turns out, eukaryotic gene regulation is remarkably more complex than the humble prokaryotic lac operon may have initially led us to believe.
So, what are the next big steps for gene regulation? In this era of epigenetics, DNA and histone modifications have been revealed as critical for the transcriptional regulation. Technical advances in imaging have already given us a near-atomic cryo-EM structure of the human Pol II PIC, and optimistically, other conformations and complexes will be unveiled through similar techniques.
An increase in the resolution of various imaging techniques has also set the stage for single-molecule imaging studies, which will be needed to understand the incredibly dynamic nature of these complexes. Future work may also hone our understanding of how three-dimensional chromatin architecture and the resulting topologically associating domains contribute to long-range enhancer contacts and gene regulation.
The cumulative efforts of many, many scientists over the last 50 years have uncovered fundamental cellular features and processes that have truly shaped our view of both genetics and cell biology. On top of that, their work has paved the way for new discoveries in the field of gene regulation, moving us closer towards a comprehensive understanding of our own cell biology.
We can’t wait to see what will come next!
<< Back to MOTIFvations Blog Home Page





