This product has been discontinued.
アクティブ・モティフの enChIP Kit®* (engineered DNA-binding molecule-mediated chromatin immunoprecipitation) は、CRISPR/Cas9 システムとChIPを組み合わせて、特定ゲノム領域に結合している複合体を単離する方法です。
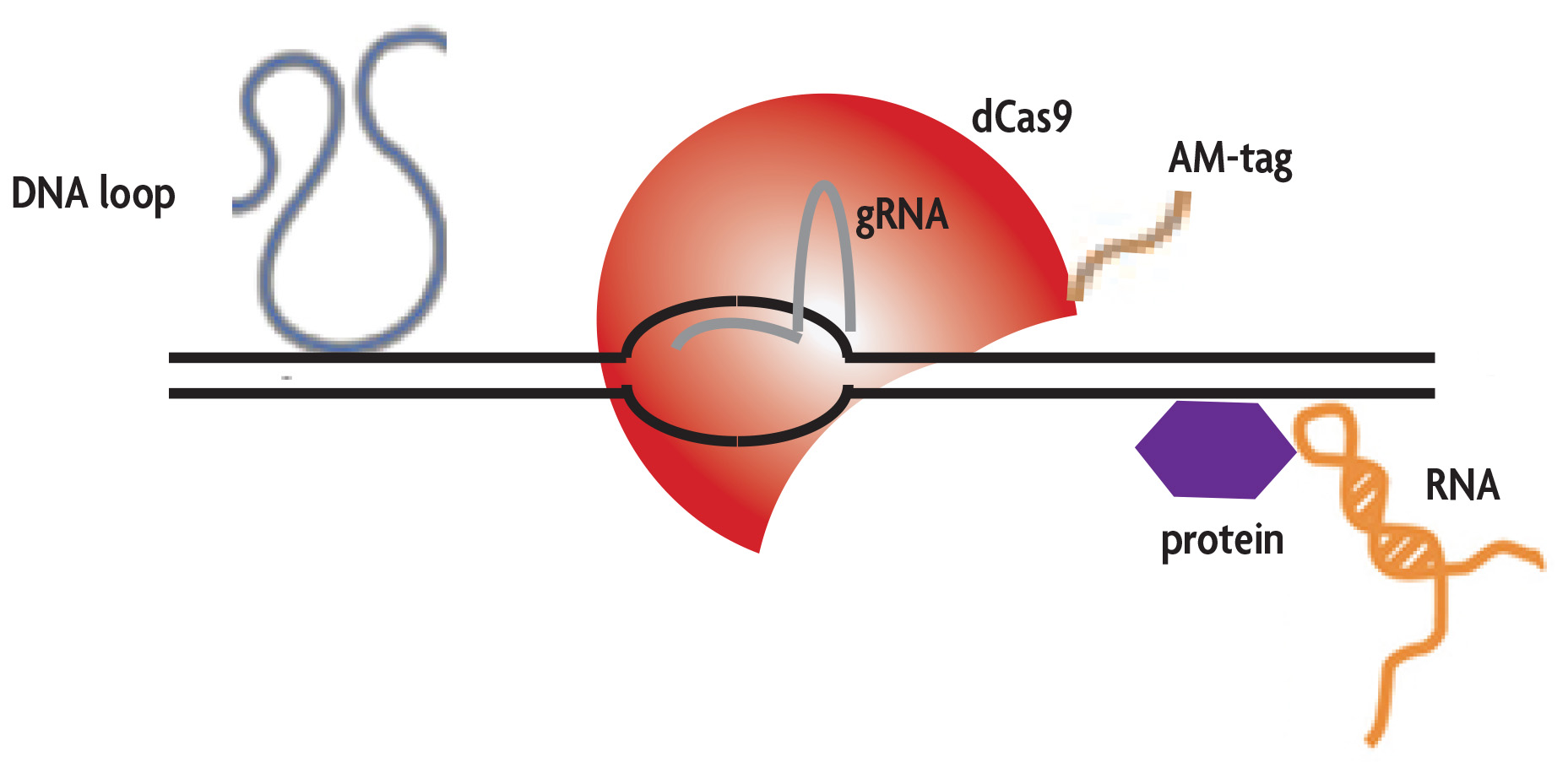
解析したいゲノム領域に対する20塩基のガイドRNA (gRNA)を設計し、タグを付加した酵素活性を失活させたCas9(dCas9)とともに、細胞に発現させます。gRNAは、dCas9と複合体を形成し、ゲノム上の特定領域に結合し、RNA-DNA複合体を形成します。タグ(AM-tag)が付加したdCas9を、抗タグ抗体を用いて免疫沈降し、回収した複合体を解析します。
enChIP Kitをお使いいただくことで、染色体ループにおけるシス/トランス制御配列の評価が行えます。さらに、CRISPR/Cas9法でのノックアウト作成において使用するgRNAのオフターゲット結合についての確認実験に使用できます。
*Patent numbers: WO2014/125668; JP5954808
enChIP® の特長
- 染色体ループにおけるシス/トランス制御配列の評価
- gRNA配列のオフターゲット結合の同定
- AM-tag/タグ抗体 を使いenChIPの濃縮効率を最適化
- オープンクロマチン領域(プロモーター、エンハンサー、インシュレーター等)の解析
enChIPに関する詳細な情報は,Method, Data, Contents タブをご覧ください。キットのマニュアル、関連する情報は、 Documents タブをご覧ください。 Resources タブからは、この技術を利用した文献情報等がご覧いただけます。
簡単な日本語資料をご用意しています。
| Name | Format | Cat No. | 価格 (税抜) | |
|---|---|---|---|---|
| enChIP® Kit | 16 rxns | 53125 | Discontinued | |
| pAM_gRNA Vector | 10 µg | 53121 | Discontinued | |
| pAM_dCas9 Vector | 10 µg | 53122 | Discontinued | |
| pAM_gRNA_CTCF Vector | 10 µg | 53123 | Discontinued | |
| pAM_dCas9_CTCF Vector | 10 µg | 53124 | Discontinued | |
How does the enChIP® Kit work?
The CRISPR/Cas system can be traced to bacteria and archaea which use the system to target and inactivate foreign genetic material. This adaptive immunity mechanism relies on acquisition of spacers, the assembly of the CRISPR RNAs and Cas proteins to the invading nucleic acids and degradation of the foreign material by nucleases1. Recently, researchers have repurposed the CRISPR/Cas9 system for genome engineering of mammalian systems2-6.
Engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) was first described in 2013 by Toshitsugu Fujita and Hodaka Fujii7,8. enChIP was designed to purify specific genomic regions using the CRISPR/Cas9 system. The targeted genomic sequence is cloned into a guide RNA (gRNA) vector in which the 5´ sequence is used to identify the DNA target sequence and the 3´ sequence forms a duplex RNA structure to bind the Cas9 protein. When the gRNA is co-expressed with a deactived (enzymatically inactive) form of the Streptococcus pyogenes Cas9 endonuclease (dCas9), specific genomic regions can be immunoprecipitated using an antibody directed against the tag. By using a catalytically inactive form of the Cas9 endonuclease, double-stranded breaks are not introduced and DNA, RNA and proteins associated with the target sequence can be recovered8-11.
In Active Motif's enChIP Kit, the dCas9 protein is expressed with Active Motif's unique AM-tag sequence, which was specifically designed for use in ChIP. The unstructured AM-tag sequence protrudes from the dCas9 protein for maximum exposure and increased immunoprecipitation efficiency. Following assembly of the gRNA and AM-tagged dCas9 to the chromatin, cells are formaldehyde fixed, lysed and the chromatin is fragmented by sonication. An antibody directed against the AM-tag is used to enrich for genomic sequences bound by the gRNA/dCas9 complex. Following immunoprecipitation, DNA can be analyzed by qPCR or NGS to identify the enriched genomic regions.
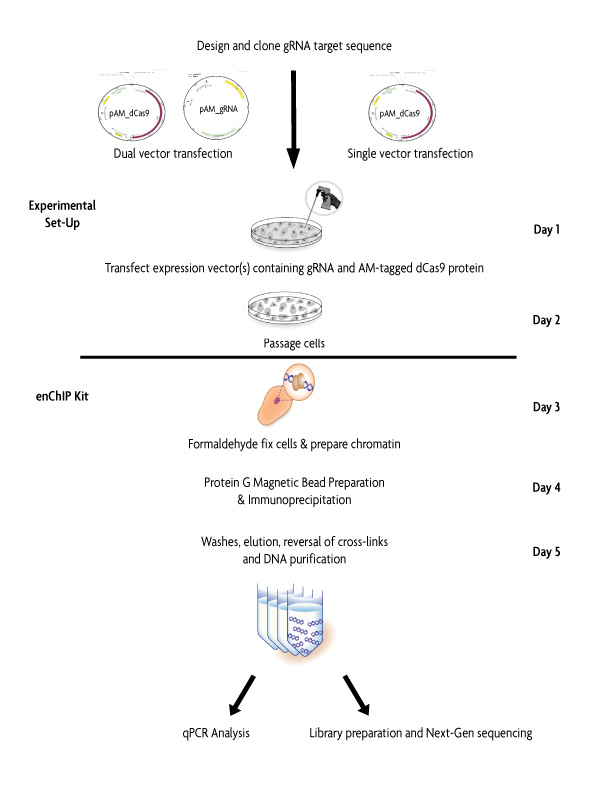
Figure 1: Flow Chart of the enChIP protocol.
Prior to using the enChIP Kit (Catalog No. 53125), a guide RNA (gRNA) target sequence is selected for a specific genomic region. The gRNA sequence is then cloned into either the pAM_gRNA expression vector for dual vector transfection, or cloned into the pAM_dCas9 expression vector for single vector transfection. 24 hours post-transfection of the gRNA and AM-tagged dCas9 expression vectors, cells are passaged. Another 24 hours later cells are formaldehyde fixed and chromatin is prepared. Protein G magnetic beads are blocked and the chromatin is pre-cleared. An overnight immunoprecipitation using the AM-Tag monoclonal antibody captures the gRNA/Cas9 genomic complexes. Following washes, the DNA is eluted from the beads, the cross-links are reversed and the enriched DNA is purified. Recovered DNA can be analyzed by qPCR or Next-Generation sequencing.
References:
- van der Oost, J. et al. (2014) Nat. Rev. Microbiol., 12(7): 479-492.
- Cong, L. et al. (2013) Science, 339(6121): 819-823.
- Mali, P. et al. (2013) Science, 339(6121): 823-826.
- Hsu, P. et al. (2014) Cell, 157(6): 1262-1278.
- Doudna, J.A. and Charpentier, E. (2014) Science, 346(6213): 1258096.
- Harrison, M.M. et al. (2014) Genes Dev., 28(17): 1859-1872.
- Fujita, T. and Fujii, H. (2013) Biochem. Biophys. Res. Commun., 439(1): 132-136.
- Fujita, T. et al. (2013) Sci. Rep., 3: 3171.
- Fujita, T. and Fujii, H. (2014) PLoS ONE, 97(7): e103084.
- Fujita, T. et al. (2015) PLoS ONE, 10(4): e0123387.
- Fujii, H. and Fujita, T. (2015) Int. J. Mol. Sci., 16(9): 21802-21812.
enChIP® Features
Identify Chromatin Looping Interactions
enChIP can reveal valuable information about potential chromosome looping interactions. gRNAs are designed to different regions of the same genomic locus. A comparison of the overlapping peak data for the various gRNAs can identify DNA sequences that repeatedly occur. These sequences may represent potential looping interaction. Additional experiments can be designed to target the possible looping site to confirm if the original genomic locus is also enriched.
enChIP ChIP-seq Results
| CTCF gRNA1 | CTCF gRNA2 | No gRNA | |
|---|---|---|---|
| Total number of reads | 39,114,618 | 40,036,875 | 38,208,604 |
| Total number of alignments | 31,486,919 | 29,584,781 | 31,765,380 |
| Unique alignments | 28,729,643 | 26,987,217 | 28,634,570 |
| Unique alignments (without duplicate reads) | 27,418,487 | 24,458,100 | 26,837,038 |
| Peak numbers | 100 | 30,822 | N/A |
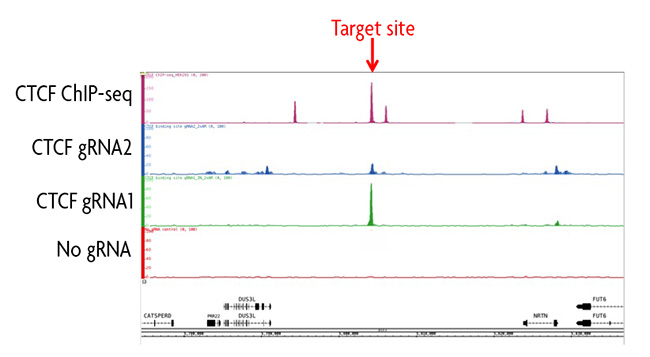
Figure 1: ChIP-seq analysis of a CTCF binding site using the enChIP Kit.
Two gRNAs were designed using CRISPRdirect targeting different sequences of a CTCF binding site on chromosome 19. Each gRNA sequence was cloned into the pAM_dCas9 Vector and transfected into HEK293T cells along with a no gRNA negative control transfection. Chromatin was prepared and immunoprecipitated according to the instructions in the enChIP Kit. Enriched DNA was analyzed by ChIP-seq and background was subtracted using the no gRNA control. Data was compared to traditional ChIP-seq data for CTCF in the same cell line. Results show gRNA1 had a strong peak at the desired target site and had little to no off-target binding events. Data for gRNA2 revealed a larger number of peaks, which represented non-specific binding of the gRNA outside of the target locus. A comparison of overlapping peak data for gRNA1, gRNA2 and the ChIP-seq data set could provide information about potential looping interactions.
gRNA Specificity
For researchers looking to perform genome editing experiments, determining the specificity of the gRNA binding site is critical. If the gRNA target sequence demonstrates off-target binding, then genome editing methods using active Cas9 protein could lead to unwanted downstream effects. Online gRNA design tools use algorithms to try and identify highly specific binding sequences, however, these tools cannot always provide accurate predictions of the in vivo response. By using Active Motif's enChIP Kit, each gRNA sequence can be tested to identify those with the most specific responses for use in genome editing applications.
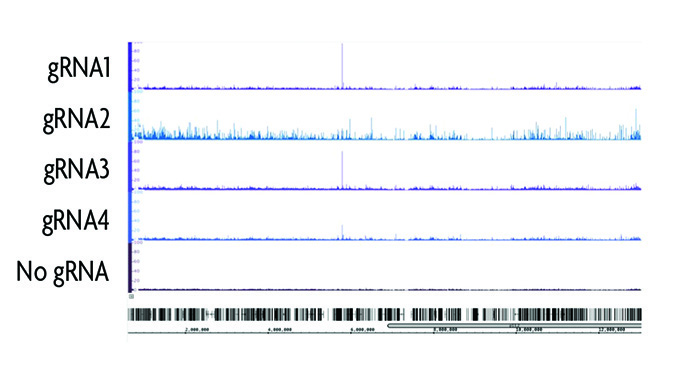
Figure 2: Determining gRNA target sequence specificity.
Four different gRNAs were designed using the CRISPRdirect program to target a 200 bp region surrounding a CTCF binding site on chromosome 19. Following transfection of the gRNA and dCas9 into HEK293T cells, chromatin was prepared and immunoprecipitated according to the instructions in the enChIP Kit. enChIP-seq results show the differences in gRNA specificity. gRNA2 has a high degree of background as compared to gRNA4 despite having only a single bp difference the target sequence. This confirms the value of performing enChIP to identify off-target binding events of the gRNA prior to use in genome engineering experiments.
Validated to Identify Promoter, Enhancer and Insulator Elements
Successful enChIP relies on the ability of the gRNA to recognize its target sequence and direct the dCas9 protein to bind the DNA. To validate the versatility of the enChIP method, Active Motif designed several gRNAs to target different genomic regions including sequences for insulator, promoter, enhancer and repressive elements. While the ChIP-seq data shows strong enrichment of the insulator, promoter and enhancer elements, the repressive region was almost undetectable. This indicates that the enChIP method is not optimized for repressive regions.
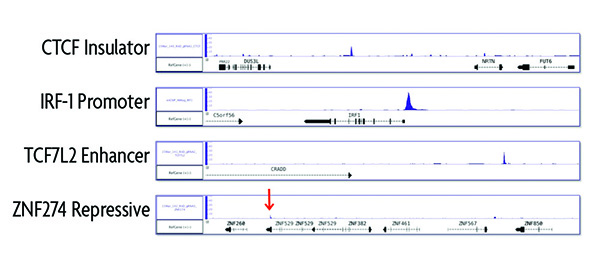
Figure 3: Comparison of enChIP performance to evaluate different genomic loci.
To evaluate different genomic loci, gRNAs were designed to target known insulator, promoter, enhancer and repressive regions of the genome. Experiments were run according to the instructions in the enChIP Kit. ChIP-seq results show highly specific enrichment of the insulator, promoter and enhancer regions. For the repressive genomic region, only a small ChIP-seq peak was observed indicating that the enChIP method is not optimized for repressive regions.
AM-tag design maximizes enChIP enrichment efficiency
Active Motif's unique AM-tag was specifically designed for use in ChIP. The AM-tag has minimal cross-reactivity with mammalian samples, thereby reducing background signal. Another benefit of the AM-tag is that the sequence is unstructured, which allows the tag to protrude from the protein of interest for maximum exposure during the immunoprecipitation for increased enrichment efficiency. The efficiency of the ChIP enrichment is shown with Active Motif's positive control pAM_gRNA_CTCF vector which was transfected into HEK293T cells and immunoprecipitated using the enChIP Kit. qPCR results show highly specific enrichment of the desired CTCF genomic locus.
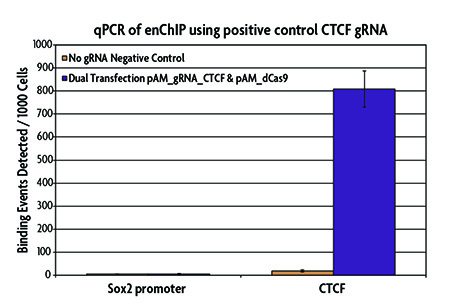
Figure 4: qPCR analysis of a positive control CTCF binding site using the enChIP Kit.
The positive control pAM_gRNA_CTCF Vector (Catalog No. 53123), which contains a gRNA sequence corresponding to a CTCF binding site on chromosome 19 (5,804,115–5,804,209), and the pAM_dCas9 (no gRNA) negative control vector were transfected into HEK293T cells. Chromatin was prepared and immunoprecipitated using the AM-tag monoclonal antibody according to the instructions in the enChIP Kit. Enriched DNA was analyzed by qPCR to evaluate a negative control (Sox2 promoter) and a positive control (CTCF) gene locus. Results show enrichment of CTCF at the expected gene locus and little to no enrichment of the no gRNA negative control or Sox2 promoter.
Contents & Storage
The enChIP® Kit (Catalog No. 53125) includes all the reagents needed to perform 16 chromatin preparation and immunoprecipitation reactions. For cloning, the pAM_gRNA Vector and pAM_dCas9 Vector are available separately. The pAM_gRNA_CTCF Vector and pAM_dCas9_CTCF Vector are also sold separately for use as an experimental control.
Please note that the enChIP Kit is shipped on dry ice and contains reagents with multiple storage temperatures inside. Please store each component at the temperature indicated below. All reagents are guaranteed stable for 6 months from date of receipt when stored properly. Do not re-freeze the Protein G Magnetic Beads after you have received this kit. This kit includes the following components:
- AM-Tag Monoclonal Antibody (1 µg/µl); Store at -20°C
- RNase A (10 µg/µl); Store at -20°C
- Proteinase K (10 µg/µl); Store at -20°C
- Carrier; Store at -20°C
- 10X PBS; Store at -20°C
- 100 mM PMSF; Store at -20°C
- Protease Inhibitor Cocktail (PIC); Store at -20°C
- Precipitation Buffer; Store at -20°C
- Anti-mouse IgG (1 µg/µl); Store at 4°C
- Fixation Buffer; Store at 4°C
- Protein G magnetic beads; Store at 4°C
- TE, pH 8.0; Store at RT
- Detergent; Store at RT
- Stop Solution; Store at RT
- Chromatin Prep Buffer; Store at RT
- ChIP Buffer; Store at RT
- Equilibration Buffer AM2; Store at RT
- Blocking Buffer AM4; Store at RT
- LS Buffer; Store at RT
- Wash Buffer AM1; Store at RT
- LiCl Buffer; Store at RT
- Elution Buffer AM4; Store at RT
- DNA Purification Binding Buffer; Store at RT
- 3 M Sodium Acetate; Store at RT
- DNA Purification Wash Buffer; Store at RT
- DNA Purification Elution Buffer; Store at RT
- DNA Purification Columns; Store at RT
pAM_gRNA Vector
Please note that the pAM_gRNA Vector is shipped at room temperature. All reagents are guaranteed stable for 6 months from date of receipt when stored properly.
- 10 µg pAM_gRNA Vector; Store at -20°C
- 250 pmol M13 Forward primer; Store at -20°C
- 250 pmol M13 Reverse primer; Store at -20°C
pAM_dCas9 Vector
Please note that the pAM_dCas9 Vector is shipped at room temperature. All reagents are guaranteed stable for 6 months from date of receipt when stored properly.
- 10 µg pAM_dCas9 Vector; Store at -20°C
- 250 pmol dCas9 Forward primer; Store at -20°C
- 250 pmol dCas9 Reverse primer; Store at -20°C
pAM_gRNA_CTCF Vector
Please note that the pAM_gRNA_CTCF Vector is shipped at room temperature. All reagents are guaranteed stable for 6 months from date of receipt when stored properly.
- 10 µg pAM_gRNA_CTCF Vector; Store at -20°C
pAM_dCas9_CTCF Vector
Please note that the pAM_dCas9_CTCF Vector is shipped at room temperature. All reagents are guaranteed stable for 6 months from date of receipt when stored properly.
- 10 µg pAM_dCas9_CTCF Vector; Store at -20°C
Resources
For more information about the enChIP technology, please view the webinar Locus-Specific Biochemical Analysis of Genome Functions Using enChIP with CRISPR presented by Dr. Hodaka Fujii.
Several online programs are available to assist with guide RNA design. A few of these programs are listed below for reference. Software instructions may change over time, so please follow the recommendations of the software provider for use.
- CRISPRdirect - http://crispr.dbcls.jp
- Michael Boutros Lab - http://www.e-crisp.org/E-CRISP/designcrispr.html
- Feng Zhang Lab - http://crispr.mit.edu
For researchers interested in lentiviral expression plasmids containing the AM-tag, these are available from Addgene, the nonprofit plasmid repository.
- pLenti_dCas9-2xAM (Plasmid #92220) https://www.addgene.org/92220/
- pLenti_dCas9-2XAM_hIRF-1 (Plasmid #92221) https://www.addgene.org/92221/
For citations using the enChIP technology, please see the publications below:
- “Identification of physical interactions between genomic regions by enChIP-Seq” by Fujita, T., Yuno, M., Suzuki, Y., Sugano, S. and Fujii, H. (2017) Genes Cells doi:10.1111/gtc.12492.
- “Isolation of Specific Genomic Regions and Identification of Their Associated Molecules by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP) Using the CRISPR System and Tal Proteins” by Hodaka Fujii and Toshitsugu Fujita (2015) Int. J. Mol. Sci 16(9), 21802-21812.
- “Identification of Proteins Associated with an IFNγ-Responsive Promoter by a Retroviral Expression System for enChIP Using CRISPR” by Hodaka Fujii and Toshitsugu Fujita (2014) PLoS ONE 9(7): e103084.