<< Back to MOTIFvations Blog Home Page
The Power of THEMIS – a cfDNA Methylome & Fragmentome Tool for Early Cancer Detection

August 16, 2024
Table of Contents:
Introduction: The Potential of Cell-free DNA in Cancer Diagnosis
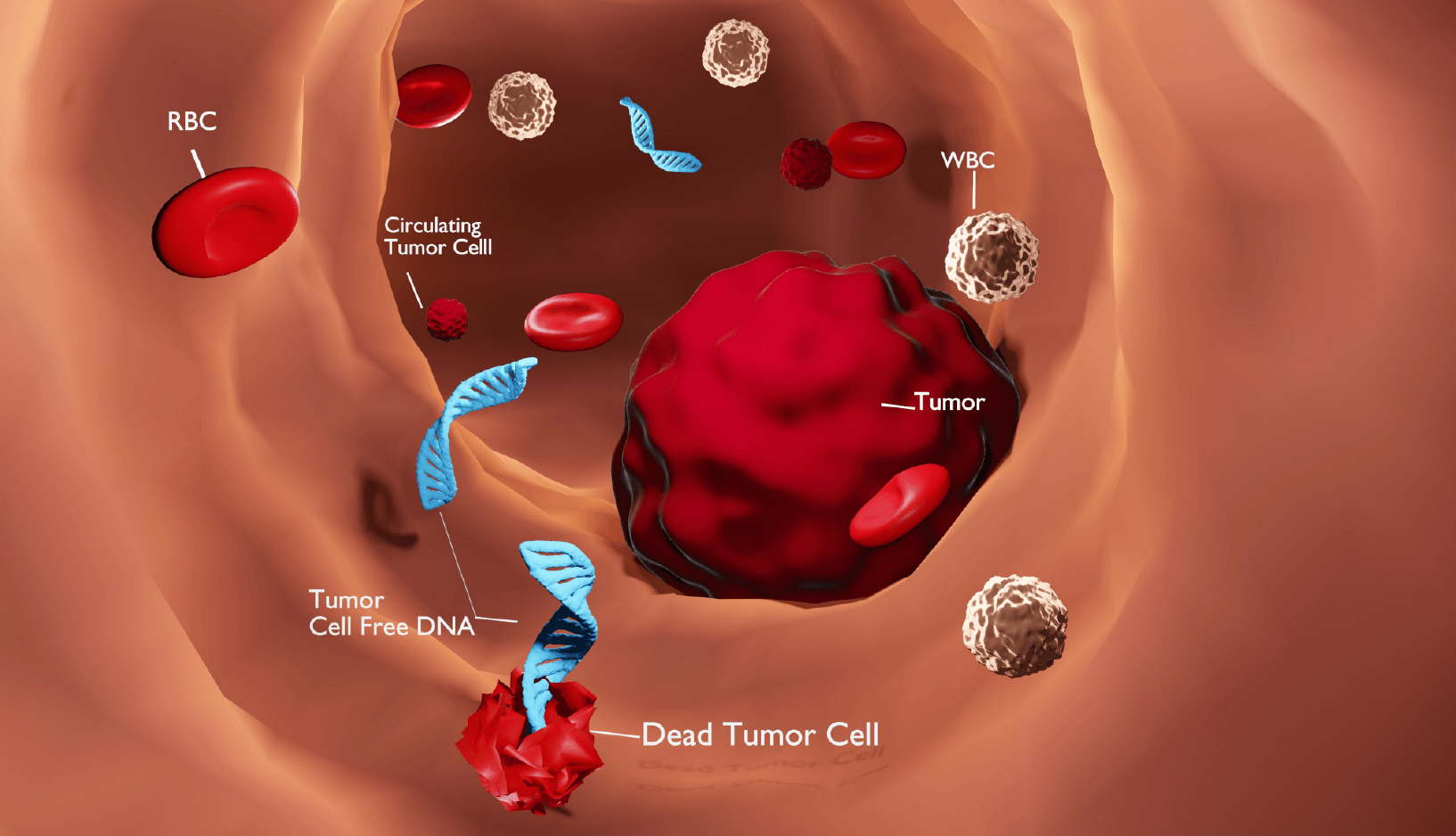
The multimodal epigenetic characterization of cell-free DNA (cfDNA) in blood plasma (from "liquid biopsies") could improve minimally invasive approaches to early cancer detection and reduce treatment morbidity/mortality. One way forward involves the integrative profiling of cfDNA methylome and fragmentome data, which remains technologically challenging; however, a recent study from Vaisvila et al. that reported the development of enzymatic methyl-seq (EM-seq) (Vaisvila et al.) supported a sensitive and non-destructive means of detecting DNA methylation (as opposed to conventional bisulfite-based methylation sequencing; Tanaka and Okamoto) while simultaneously evaluating fragmentation.
News of this newly developed protocol prompted researchers guided by Fengwei Tan and Shugeng Gao (Chinese Academy of Medical Sciences and Peking Union Medical College) to adapt EM-seq to analyze the small amounts of cfDNA generally obtained from the blood plasma of cancer patients. In their new study in Nature Communications (Bie and Wange et al.), the Tan and Gao groups now report how the integration of crucial tumor genetic and epigenetic characteristics defined through the analysis of cfDNA supported the development and validation of "THEMIS" or "THorough Epigenetic Marker Integration Solution" as a multicancer detection test that facilitates sensitive and accurate early detection.
Integrating the Analysis of Cell-free DNA methylation, Fragmentation, and Copy Number Alterations into THEMIS
To develop THEMIS, Bie and Wange et al. applied shallow whole-methylome sequencing to plasma samples isolated during a multicenter, case-control, observational MONITOR (Multi-Omics Noninvasive Inspection of TumOr Risk) study that involved seven common cancer types (780 patients) and healthy controls (497 patients). As the mild enzymatic reactions minimized DNA damage during the analysis of cfDNA methylation, the team could integrate the simultaneous profiling of multiple types of cancer genomic features - fragmentation and copy number alteration - to enhance cancer detection. They also developed computational methods that extracted DNA methylation (Allen Chan et al.), fragment size (Cristiano et al.), copy number alterations (Leary et al.), and fragment end motif (Jiang et al.) cancer-associated features from the plasma cfDNA genome to then construct an ensemble machine learning classifier (THEMIS) that distinguished cancer patients from healthy controls with high sensitivity.
Just What Can EM-seq and THEMIS Do?
The team first analyzed associations among fragmentation, methylation, and copy number alteration profiles in the plasma cfDNA genome of healthy controls and colorectal cancer samples to investigate whether multimodal genomic features generate complementary signals in cancer detection. Overall, the modest-to-weak associations between multimodal genomic features suggest that they provide concordant and complementary information, implying that integrative analyses may enhance the detection power of cancer-associated signals.
The team next applied whole-methylome sequencing via EM-seq to plasma cfDNA samples from all cancer patients and healthy controls of the MONITOR study to develop and validate computational approaches for multicancer detection. Encouragingly, data on each detected feature separated cancer patients from healthy controls, supporting their utility in cancer detection. While late-stage samples displayed more apparent cancer-type clustering than early-stage samples (which suffer from lower levels of cfDNA), the data still separated early-stage cancers from healthy controls.
The authors subsequently applied machine learning methods to the cfDNA-derived data to increase the sensitivity of cancer detection, capture weak tumor signals more sensitively, and avoid problems related to technical and biological noise, and then constructed the ensemble THEMIS classifier to boost the performance of their multicancer detection model. Encouragingly, the application of THEMIS reduced the number of healthy individuals misclassified as cancer patients and increased the number of correctly identified cancer patients, which confirmed the author's hypothesis that integrative analysis of multimodal cancer genomic features would improve the detection specificity and sensitivity over profiling individual features alone. THEMIS consistently and accurately detected seven cancer types in early-stage cancer patients, achieving high sensitivity across disease types and stages; however, prediction score and detection sensitivity increased with increasing clinical stage (as occurred for individual modalities).
Finally, and perhaps most interestingly, the team also profiled plasma cfDNA methylation and fragmentation patterns at cancer tissue-specific accessible chromatin regions, which allowed the accurate localization of the tissue origin of cancer signals, which represents a crucial component of a multicancer test to guide follow-up diagnostic workups.
The Power and the Limitations of THEMIS
Overall, this proof-of-concept study defines the power of analyzing multimodal genetic and epigenetic features of cfDNA in liquid biopsies in improving early detection and localization of multiple forms of cancer; indeed, the advantages of THEMIS over currently available imaging-based diagnostic procedures highlight the potential real-world clinical applications of this approach. Overall, the authors highlight the potential of THEMIS in the general screening of populations with low to average cancer risk.
For more on how integrating the cfDNA methylome and fragmentome may boost cancer detection, see Nature Communications, September 2023.
About the author

Stuart P. Atkinson, Ph.D.
Stuart was born and grew up in the idyllic town of Lanark (Scotland). He later studied biochemistry at the University of Strathclyde in Glasgow (Scotland) before gaining his Ph.D. in medical oncology; his thesis described the epigenetic regulation of the telomerase gene promoters in cancer cells. Following Post-doctoral stays in Newcastle (England) and Valencia (Spain) where his varied research aims included the exploration of epigenetics in embryonic and induced pluripotent stem cells, Stuart moved into project management and scientific writing/editing where his current interests include polymer chemistry, cancer research, regenerative medicine, and epigenetics. While not glued to his laptop, Stuart enjoys exploring the Spanish mountains and coastlines (and everywhere in between) and the food and drink that it provides!
Contact Stuart on Twitter with any questions
Related Articles
An Epigenomic Compendium of Sporadic ALS Cases – R-loops & the DNA Methylome
February 22, 2024
Researchers report a compendium that describes the involvement of genetic variants and alterations to DNA methylomes and R-loop accumulation in sporadic cases of amyotrophic lateral sclerosis. Sounds daunting, but we break down some key concepts that show, once again, the relevance of epigenetic analysis in human disease.
Read More
LINE-1 Transposable Elements: Can they Provide Liquid Biopsy Biomarkers for Early Cancer Detection and Management?
April 4, 2023
Detection of LINE-1 ORF1p in liquid biopsies has been proposed as an effective means of diagnosing cancer early and improving therapeutic outcomes. In this article, we take a look at two related, innovative studies describing how LINE-1 transposable elements may provide new biopsy biomarkers whose detection will improve the early diagnosis and long-term management of many cancers.
Read More
Epigenomic Analysis of Cancer Liquid Biopsies - The Power and Limitations
January 17, 2024
The straightforward epigenomic analyses of liquid biopsies from cancer patients represent a minimally invasive means of understanding tumor gene expression programs and supporting patient management. Read how cfDNA as a sample type and new approaches including cfChIP-Seq and cfMeDIP-Seq are being explored for disease profiling.
Read More
DNA Methylation’s Impact on cfDNA Fragmentation – Considerations When Choosing Biomarkers
August 29, 2023
A new understanding of how differential DNA methylation may impact cell-free DNA fragmentation may help in the identification of powerful biomarkers in liquid biopsies.
Read More
<< Back to MOTIFvations Blog Home Page