Fixed ATAC-Seq Services Overview
Fixed ATAC-Seq from Active Motif is ideal when working with precious, low-input, or difficult-to-process samples that require stabilization prior to chromatin accessibility analysis. This method allows you to fix cells with formaldehyde immediately after collection, preserving the native chromatin state and enabling batch processing or time-course studies with minimal variability. Fixed ATAC-Seq is especially useful for primary cells, research samples, or experiments where immediate processing or cryopreservation isn't feasible.
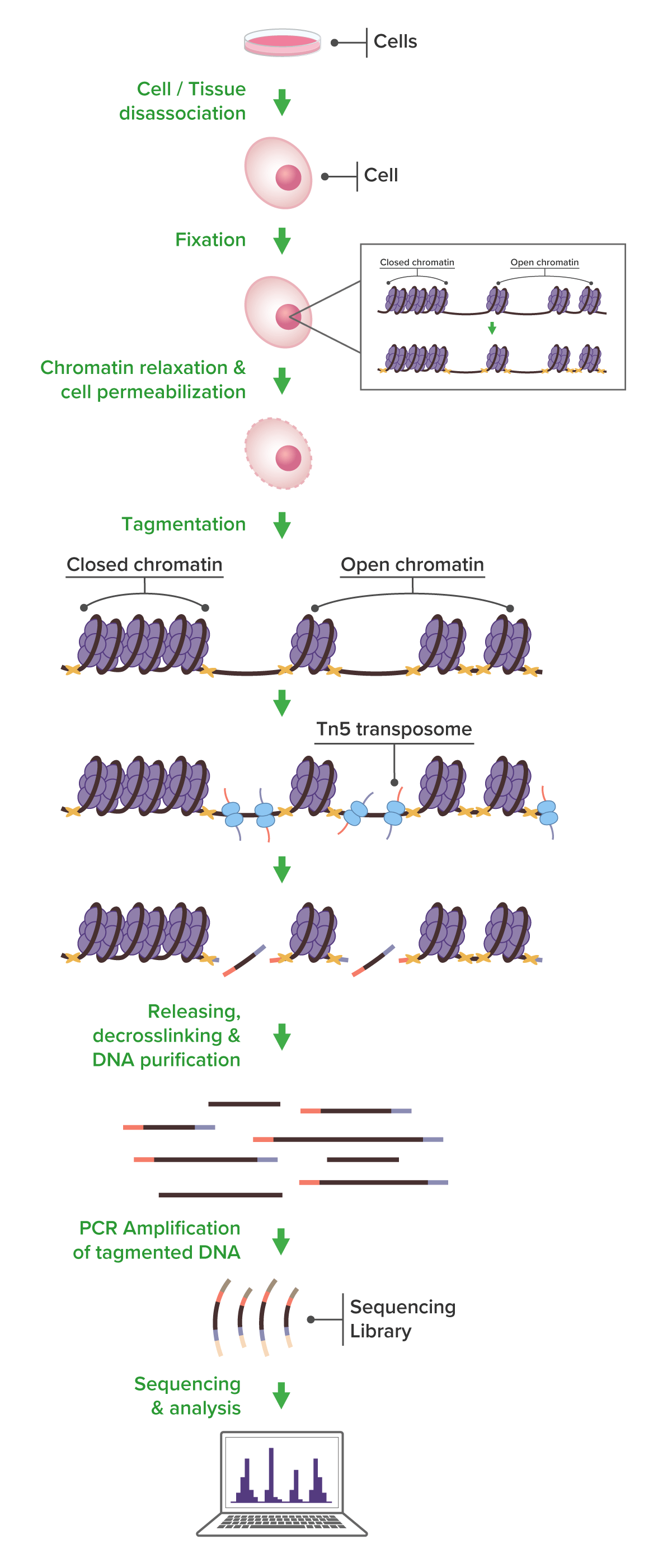
The Fixed ATAC-Seq process begins with fixation of intact whole cells using 1% formaldehyde to preserve chromatin structure. After fixation, nuclei are isolated and subjected to transposition using a hyperactive Tn5 transposase, which simultaneously fragments accessible regions of the genome and tags them with sequencing adapters. After decrosslinking and purification, the resulting libraries are then amplified and sequenced using next-generation sequencing (NGS). This approach maintains chromatin integrity while allowing flexible sample handling and batching, making it well-suited for time-course experiments and high-throughput studies. Our end-to-end service includes comprehensive data analysis support.
Let our team of ATAC-Seq experts handle the entire workflow, from sample prep to bioinformatics. With optimized protocols and in-depth analysis included, we make it easy to generate reliable, publication-ready chromatin accessibility data.
Fixed ATAC-Seq Highlights:
- Compatible with formaldehyde-fixed samples.
- Enables efficient batch processing and consistent results across time-course and drug-treatment experiments.
- Optimized for primary cells.
- End-to-end service.
- Fast turnaround time.
Active Motif’s End-to-End Fixed ATAC-Seq Service includes:
- Chromatin relaxation and cell permeabilization
- Tagmentation using Tn5 transposase for library generation
- DNA Release, decrosslinking and purification
- PCR amplification
- Next-generation sequencing
- Comprehensive bioinformatics and data analysis
- Delivery of publication-ready figures
Fixed ATAC-Seq Services Data
(Click images to enlarge)
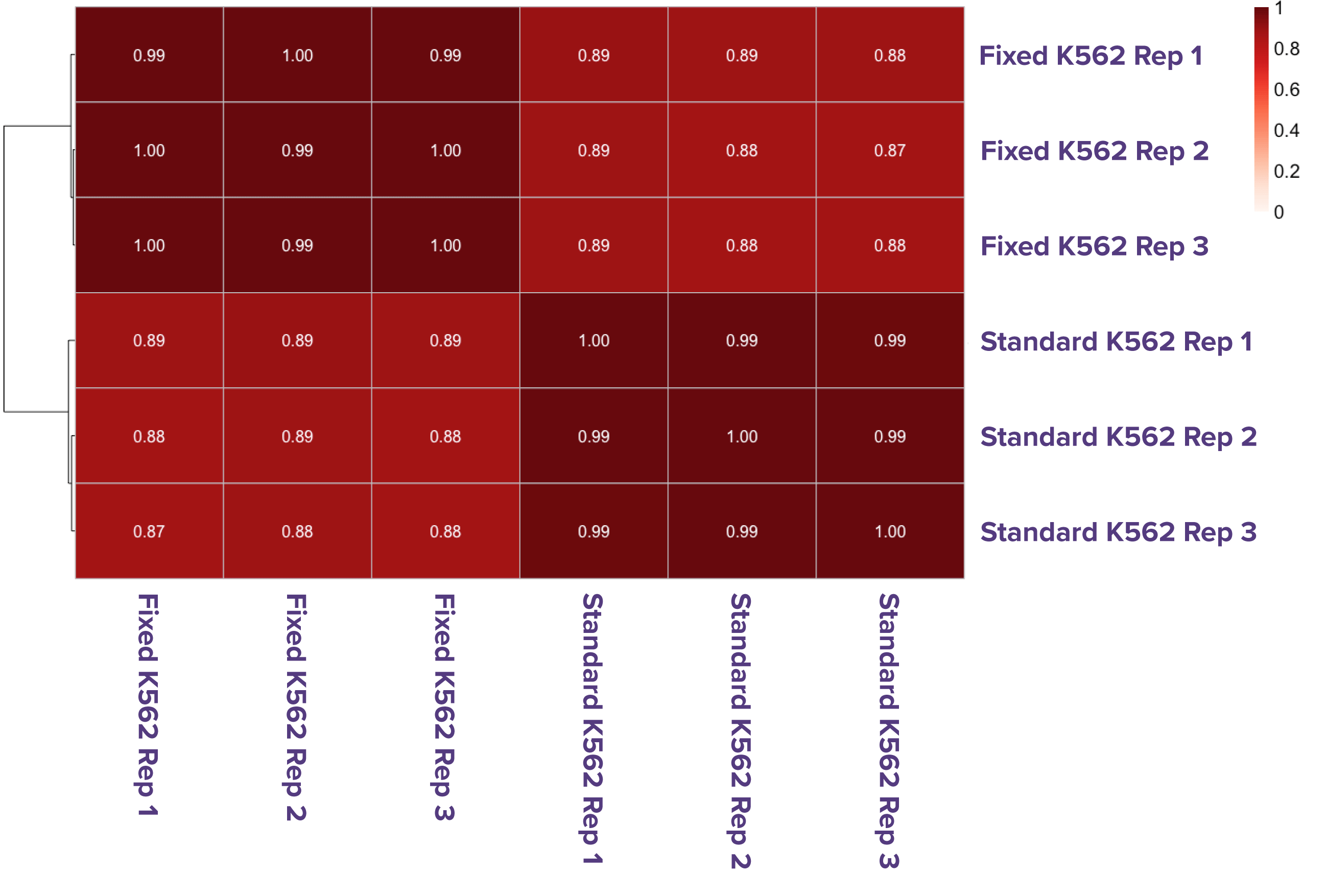
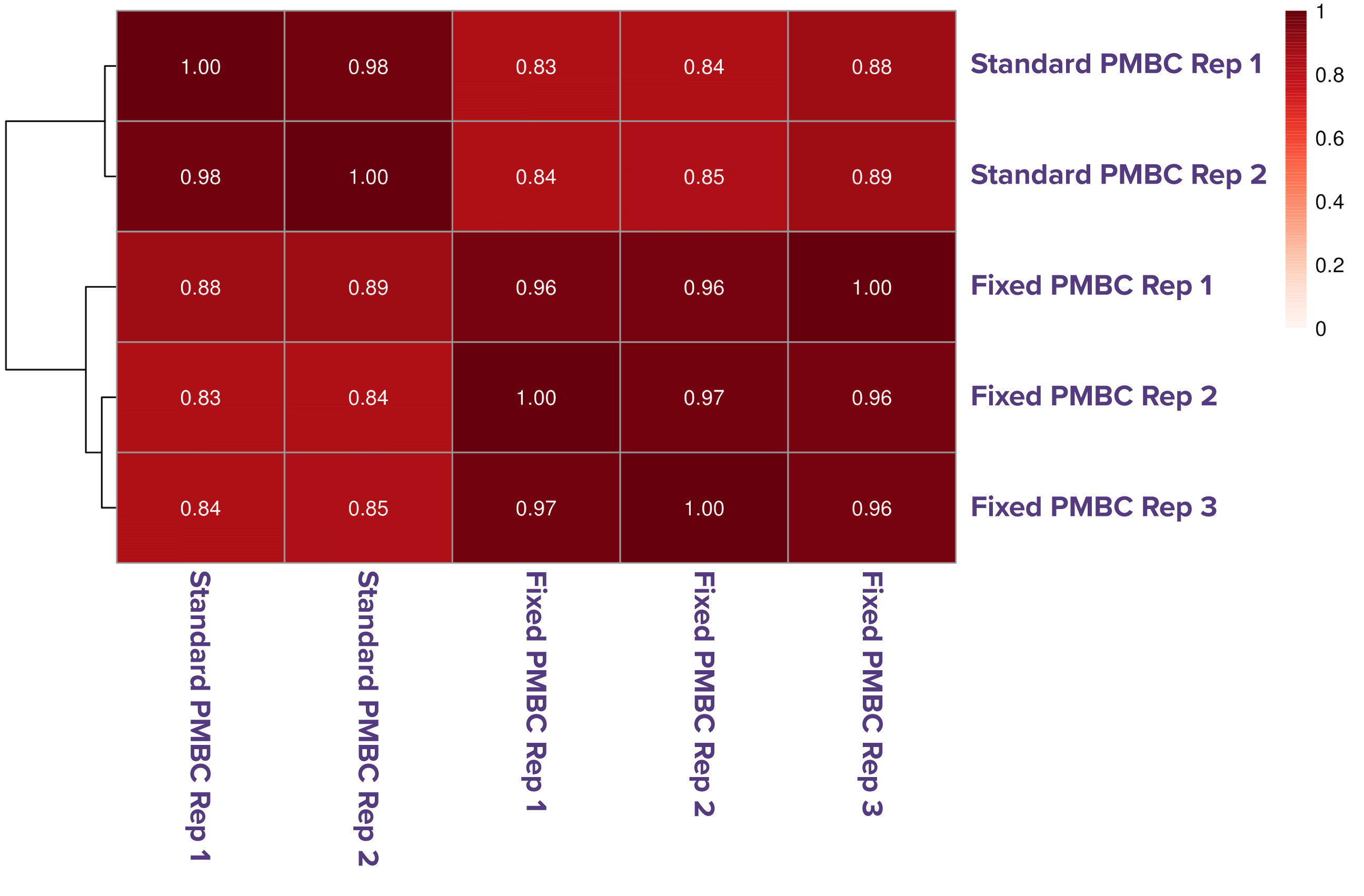
Figure 1. High correlation between Native and Fixed ATAC-Seq in both K562 cells and PBMCs.
ATAC-Seq data generated using the standard (native) and Fixed ATAC-Seq protocols show strong concordance in both human erythroleukemic (K562) cells and primary PBMCs. K562 cells (100K) were processed in triplicate with each method, demonstrating high correlation between native and fixed conditions (A). Similarly, 100K PBMCs processed in duplicate (native) and triplicate (fixed) also show consistent, high-quality data across methods.
(Click images to enlarge)
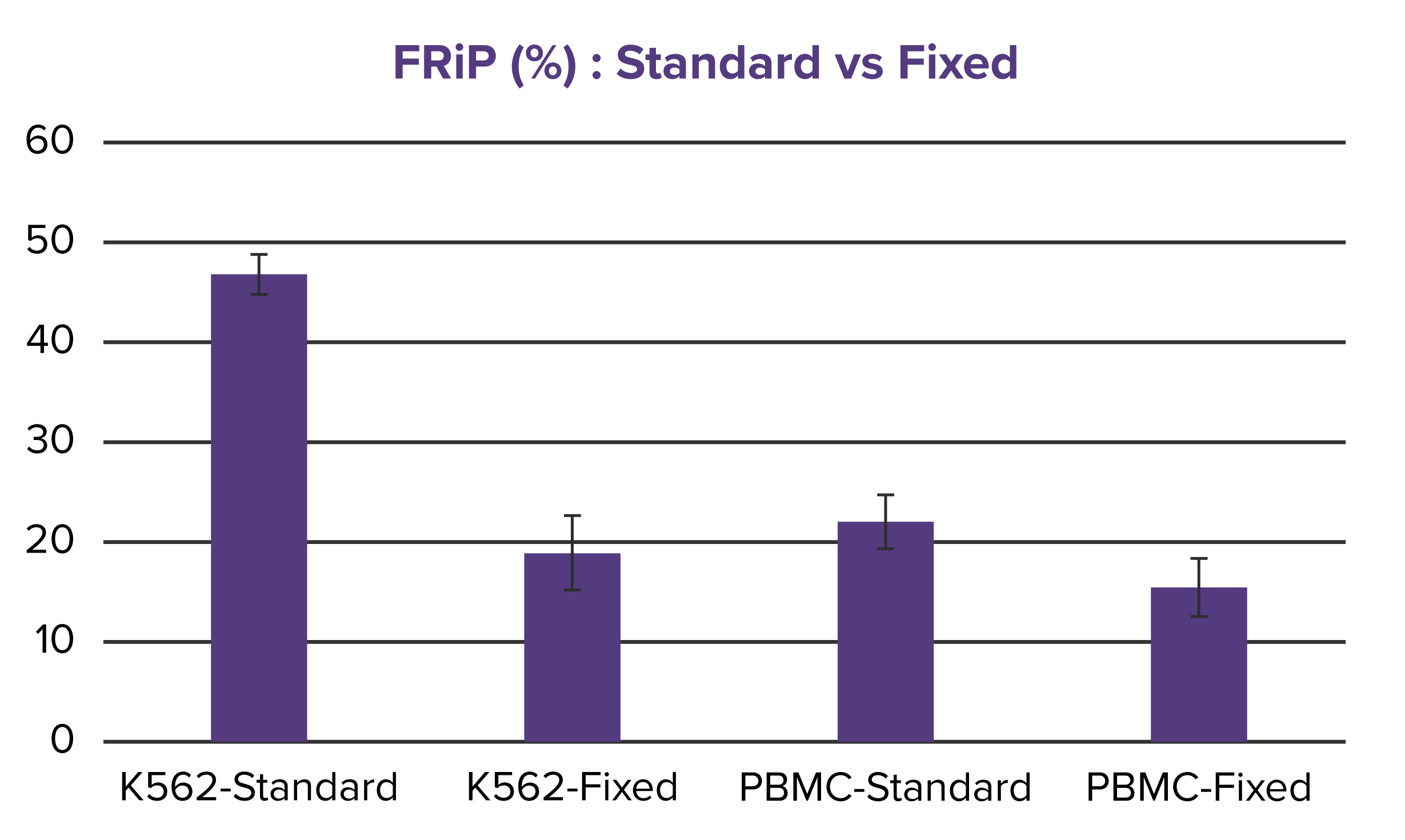
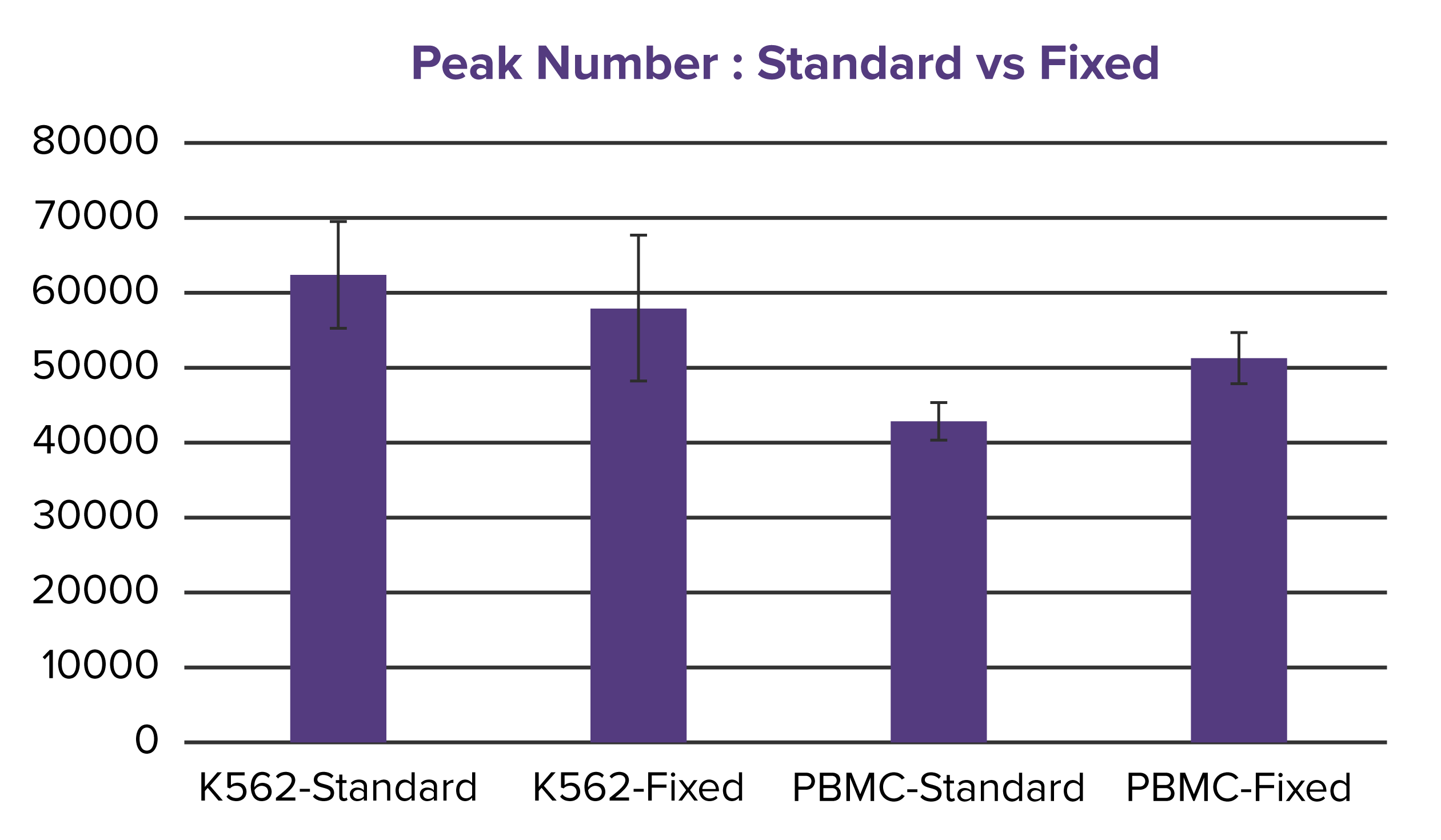
Figure 2. Comparable quality metrics between Native and Fixed ATAC-Seq.
FRiP scores (a) and peak counts (b) are shown for both Native and Fixed ATAC-Seq performed on K562 cells and primary PBMCs. While a decrease in FRiP score is observed in Fixed K562 samples compared to Native ATAC-Seq, the number of peaks remains comparable. In PBMCs, both FRiP scores and peak numbers are consistent across methods, with FRiP values aligning with expectations for primary cells (~25%).
(Click images to enlarge)
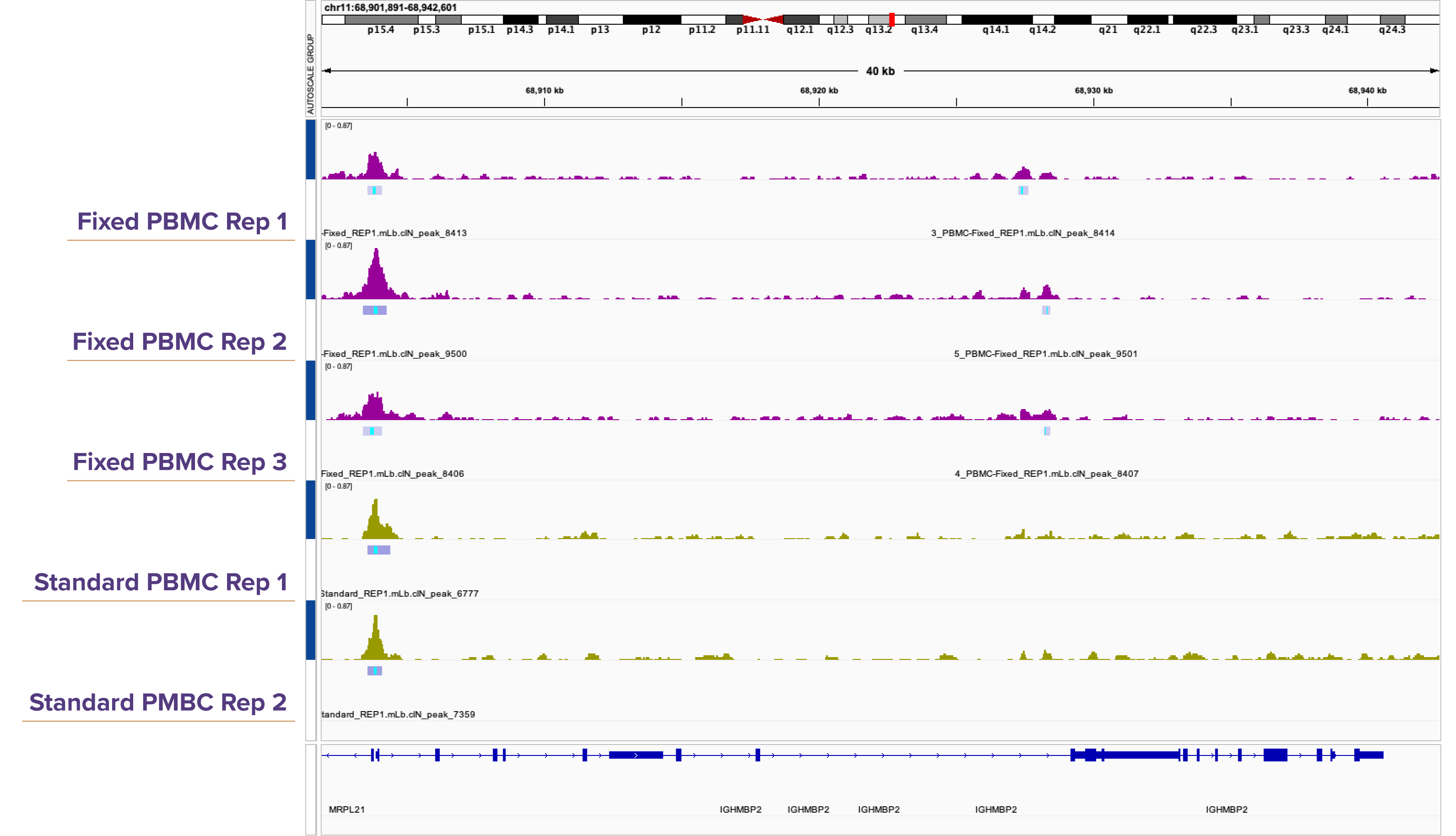
Figure 3. Genome browser tracks demonstrate high reproducibility between Native and Fixed ATAC-Seq in PBMCs.
Genome browser views highlight consistent peak calling between Native and Fixed ATAC-Seq in primary PBMCs. The tracks show strong agreement in signal profiles, with both methods identifying the same accessible chromatin regions.
 (Click image to enlarge)
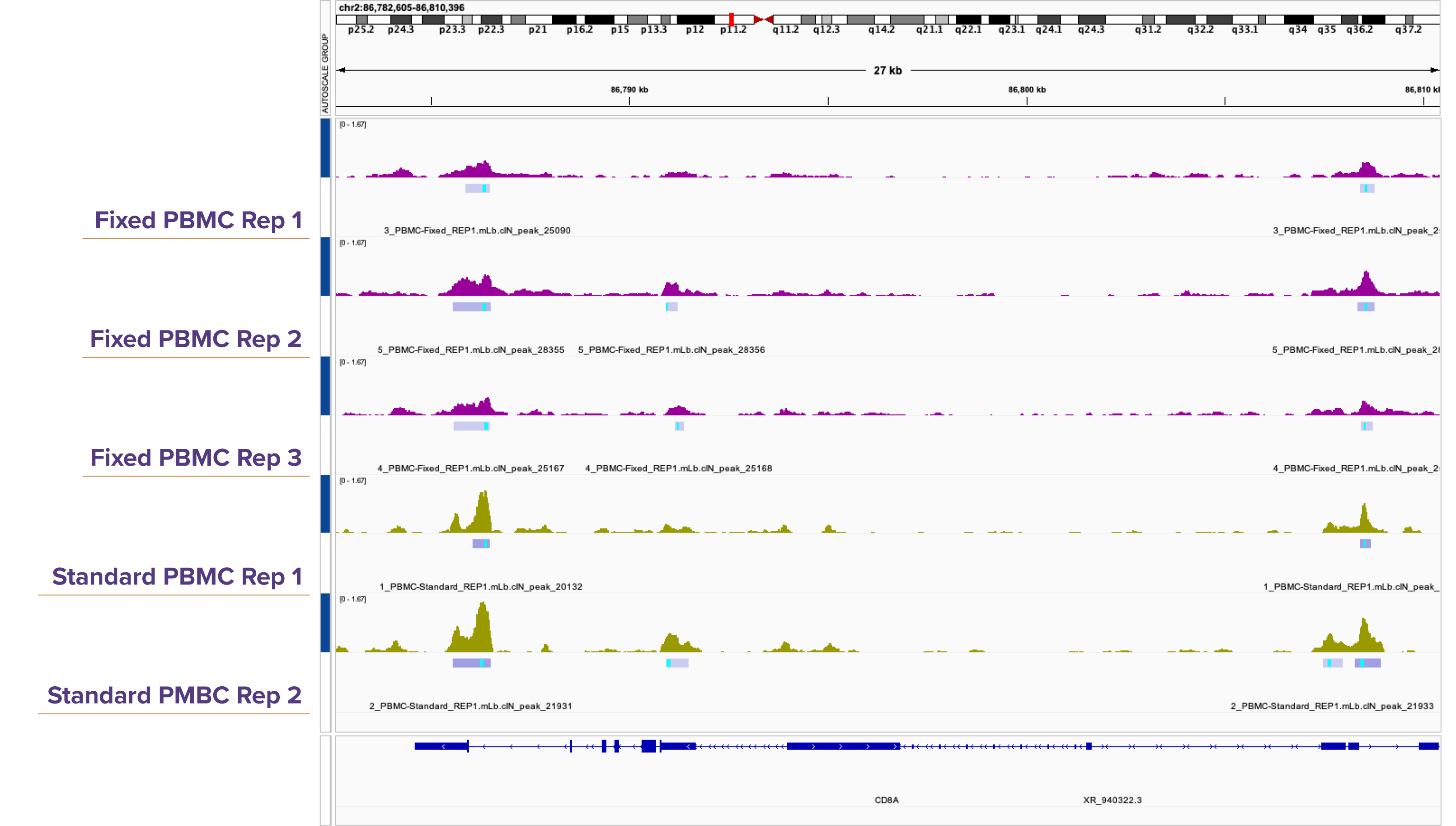
(Click image to enlarge)Figure 4. Genome browser tracks demonstrate high reproducibility between Native and Fixed ATAC-Seq in K562.
Both methods reveal highly similar peak patterns, with strong alignment in location, confirming reproducible detection of open chromatin regions.
Fixed ATAC-Seq Services FAQs
Can I expect to obtain the same quality data from Fixed ATAC-Seq as from regular ATAC-Seq?
Can I fix samples using lower concentrations of formaldehyde?
Can I submit FFPE samples for Fixed ATAC-Seq Service?
Can I fix isolated nuclei instead of whole cells for Fixed ATAC-Seq?
Fixed ATAC-Seq Services Documents
Fixed ATAC-Seq Services Sample Submission Portal
Our online sample submission portal allows you to easily upload your service project samples and track your project status. Follow the sample submission instructions in the portal to ensure that all your samples arrive at Active Motif in the best possible condition and properly associated with your project.
You might also be interested in:
- Fixed-Cell ATAC-Seq Kit
- ATAC-Seq Service
- Single-Cell ATAC-Seq Service
- [Blog] Exploring ALS (Amyotrophic Lateral Sclerosis) with iPSCs and ATAC-Seq
- [Blog] Complete Guide to Understanding and Using ATAC-Seq
- [Podcast] ATAC-Seq, scATAC-Seq and Chromatin Dynamics in Single-Cells (Jason Buenrostro)
- [Webinar] ATAC-Seq: Mapping Open Chromatin
- [Video] Intro to ATAC-Seq: Method overview and comparison to ChIP-Seq
| Name | Cat No. | 価格 (税抜) | |
|---|---|---|---|
| Fixed ATAC-Seq Service | 25204 | Get Quote |